Abstract
Background
Hepatitis B virus (HBV)-encoded X antigen, HBx, assists in the development of hepatocellular carcinoma (HCC) through complex mechanisms. Our results provide new insights into the EZH2 epigenetic repression of let-7c that promotes HCC migration induced by HBx. Thus, let-7c and HMGA2 represent key diagnostic markers and potential therapeutic targets for the treatment of HBV-related HCC.
Results
We investigated the epigenetic regulation of let-7c, an important representative miRNA in liver tumor metastasis, in human HCC cells to verify the effect of HBx. Based on quantitative PCR (qPCR) of mRNA isolated from tumor and adjacent non-tumor liver tissues of 24 patients with HBV-related HCC, EZH2 expression was significantly overexpressed in most HCC tissues (87.5%). We executed a miRNA microarray analysis in paired HBV-related HCC tumor and adjacent non-tumorous liver tissue from six of these patients and identified let-7c, miR-199a-3p, and miR-99a as being downregulated in the tumor tissue. Real-time PCR analysis verified significant downregulation of let-7c and miR-99a in both HepG2X and Hep3BX cells, which stably overexpress HBx, relative to parental cells. HBX enhanced EZH2 expression and attenuated let-7c expression to induce HMGA2 expression in the HCC cells. Knockdown of HMGA2 significantly downregulated the metastatic potential of HCC cells induced by HBx.
Conclusions
The deregulation of let-7c expression by HBx may indicate a potential novel pathway through deregulating cell metastasis and imply that HMGA2 might be used as a new prognostic marker and/or as an effective therapeutic target for HCC.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is the seventh most general cancer in the world and the third leading origin of death among cancers worldwide. And, the highest incidence in the world has arisen in Asia and Africa [1]. In particular, hepatitis B virus (HBV) infection is the major factor in chronic liver disease and HCC. Therefore, HBV infection remains a global health issue with eventful morbidity and mortality [2,3,4] The HBV X protein (HBx) is an oncogenic protein that is required for infection and replication of the virus and is closely related to the development of HCC. Therefore, the HBx is considered to take a pivotal part in the molecular pathogenesis of HBV-correlative HCC [5,6,7]. HBx promotes migration and invasion of HCC cells through upregulating osteofibronectin and matrix metalloproteinases to reduce intercellular adhesion [8, 9]. Multiple reports have verified that HBx can operate numerous cancer-related genes, and pilot the formation and progression of tumors [10,11,12,13]. Hence, HBx has an essential position during HCC progression resulting in HBV chronic infection [14, 15]. However, the complex mechanisms of HBX regulating tumor development are still not well known.
Enhancer of zeste homolog 2 (EZH2) is a catalytic subunit of the Polycomb Repressor Complex 2 (PRC2), having the form of a complex with SUZ12 and EED protein. This complex catalyzes the trimethylation of histone H3 lysine 27 (H3K27me3), which restrains the transcription of many genes. The function of EZH2 participates in a heavy role in the regulation of genes during biological development, especially in cancer, where abnormal performance is often observed [16, 17]. During the past decade, EZH2 has been identified as an oncogene in many types of cancers and acts through epigenetic repression of various tumour suppressor genes [18,19,20,21,22,23]. Besides, inhibiting the transcription of many genes, EZH2 has also been presented to suppress the expression of many tumor suppressor microRNAs (miRNAs), such as miR-125a, miR-125b, miR-138-5p, and miR-320c in HCC, thus assisting HCC tumorigenicity and metastasis [24,25,26,27].
In this study, we wanted to understand the relationship between the molecular mechanism of between HBx and EZH2 in promoting HCC. In addition, we addressed the functional interplay between HBx and the miRNA machinery to identify potential initiating events for HCC metastasis and to identify the mechanism by which HBx and EZH2 regulate these genes.
Results
EZH2 is frequently overexpressed in HCC patients and correlated with overall survival
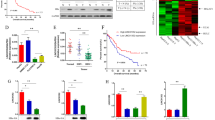
We verified the expression of EZH2 in 24 pairs of HBV-related HCC tumor tissues compared to normal adjacent liver tissue. This study assessed the levels of EZH2 mRNA through qPCR. The comparative levels of EZH2 in the tumor section and the normal tissues (n = 24) are shown in Fig. 1A. In most patients (21/24, 87.5%), EZH2 was upregulated in HCC tissues relative to normal adjacent tissues, with a mean ± SEM fold increase of 7.01 ± 1.39 (P < 0.001, Fig. 1B). The EZH2 levels in 20 of 24 tumor tissue samples (83.3%) were increased by ≥ 2-fold in contrast to in normal adjacent tissues. Futhermore, in order to investigate the clinical impact of EZH2 on HCC progression, we also assessed the relationship between cellular levels of EZH2 mRNA and the overall suvival in HCC. HCC patients with higher EZH2 expression compared to the normal control had significant results and high level of EZH2 indicated poor suvival (Fig. 1C, D). Based on the above data, we speculated that the overexpression of EZH2 may be related to HBV infection.
Analysis of EZH2 expression in HCC tumor samples. A. EZH2 mRNA relative to actin (control) was measured by qRT-PCR in 24 HCC tumor tissue samples in contrast to adjacent normal liver tissue from the same patients. B. The mean ± SEM of the relative EZH2 mRNA level for all 24 cases. C. The level of EZH2 between HCC tumors and normal control. D. The overall survival of EZH2 in HCC patients from TCGA database. (*p < 0.05, ***p < 0.001)
HBx upregulates EZH2 expression
To determine whether the proportion of EZH2 is correlative to HBx overexpression, we used an encoding HBx plasmid into HepG2 and Hep3B cells on two human HCC cell lines. The expression of EZH2 was obviously elevated in HBx-expressing HepG2 and Hep3B cells as compared with the empty vector (Fig. 2A). In addition, we applied HepG2X and Hep3BX, which stably overexpress HBx[10, 11], to determine the proportion of HBx in contrast to their parental cell lines HepG2 and Hep3B, separately. EZH2 mRNA levels were heightened in HepG2X and Hep3BX cells as compared with the parental cells (Fig. 2B). Similarly, the level of EZH2 protein was also raised in HepG2X and Hep3BX cells (Fig. 2C). The results provided evidence that HBx upregulates EZH2 expression in HCC cell lines.
Upregulation of EZH2 proportion by HBx. A. HepG2 and Hep3B cells were temporarily transfected with a control vector plasmid (Ctl) or an HBx expression plasmid at the indicated concentration. EZH2 expression was surveyed by qRT-PCR. B. RT-PCR analysis of EZH2 expression in HepG2X (G2X) and Hep3BX (3BX) cells in contrast to parental cells HepG2 (G2) and Hep3B (3B), respectively. C. Western blot analysis of EZH2 protein in HepG2X and Hep3BX cells in contrast to parental cells HepG2 and Hep3B. The GAPDH and α-tubulin as controls
let-7c and miR-99a are key regulatory miRNAs in HBV-related HCC
To verify the influence of HBx on miRNA representation, we examined miRNA levels in HCC tissues gathered from six patients in contrast to HBV-related HCC tumors with non-tumor tissue samples. Seventy-two miRNAs were found to present remarkable changes in expression in all six paired biological samples (Fig. 3A). Only three miRNAs were downregulated in all six HCC tissue samples, let-7c, miR-99a, and miR-199a-3p (Fig. 3B). To identify whether these regulated miRNAs were thought of together with HBx in HCC cells, we verified their relative levels of expression in HepG2X and Hep3BX cells through qRT-PCR. let-7c and miR-99a levels were lower in the rates of HepG2X and Hep3BX cells relative to the parental cells (Fig. 3C, D). Besides, the level of miRNA-199a-3p was decreased only in HepG2X cells as compared with the parental cells. Simultaneously, a tricistronic miRNA cluster bearing miR-99a/let-7c/miR-125b-2 resides on hsa21. Moreover, to investigate the clinical impact of let-7 C, mir-199a-3p, and mir-99a on HCC progression. The overall suvival of these miRNAs had been evaluated with the KM plotter and the resulut indicated that lower let-7c and mir-99a have poor survial in HCC patients (Fig. 3E). These consequences supported an opinion that HBx downregulates the expression of let-7c and miR-99a.
The profile of miRNAs downregulated by HBx. A. Heatmap of 72 miRNAs that were differentially expressed in six HBV-related HCC tumors (case number 3, 13, 48, 58, 60, 68) as compared with their non-tumor tissue controls. qRT-PCR analysis of these three miRNAs was performed B in the HCC tumor (T) and non-tumor (N) tissue samples and C in HepG2X and D in Hep3BX cells as compared with the parental cells HepG2 (G2) and Hep3B (3B), respectively. E. The overall survival of let-7c, mir-199a-3p, and mir-99a in HCC patients from TCGA database
EZH2 represses let-7c and miR-99a expression
For the purpose of determining whether the expression of microRNAs is repressed because of EZH2 overexpression, we utilized a plasmid encoding EZH2 into HepG2 and Hep3B cells. The level of EZH2 was considerably elevated in EZH2-expressing HepG2 and Hep3B cells in contrast to the empty vector (Fig. 4A). To investigate the phenomenon of EZH2 knockdown, we executed short hairpin RNA (shRNA) experiments in HepG2X cell lines. For this purpose, we applied shRNAs that targeted positions in the EZH2 mRNA nucleotide sequence. Consistent with the results, let-7c and miR-99a expression were significantly increased in EZH2 knockdown HepG2X cells relative to the parental cells (Fig. 4B). let-7c evidenced a greater fold increase than miR-99a in EZH2 knockdown cells. An inhibitor of S-adenosylhomocysteine hydrolase, DZNep, leads to the maintenance of S-adenosylhomocysteine, accordingly restraining S-adenosyl-L-methionine dependent methyltransferases as well as EZH2 [28, 29]. DZNep also reportedly depletes EZH2 protein [30]. As expected, HCC cells handled with DZNep (10 µM) declared a substantial decline in EZH2 expression and showed a corresponding significant increase in let-7c and miR-99a representation (Fig. 4C, D). These results identified that DZNep enhances let-7c and miR-99a representation in HepG2X and Hep3BX cells.
Analysis of EZH2 repression of let-7c and miR-99a expression. A. EZH2 was transfected into HepG2 and Hep3B cells. The expression of EZH2 was elevated in EZH2-expressing HepG2 and Hep3B cells as compared with cells transfected with the empty vector. let-7c and miR-99a expression was normalized against the control miRNA U48, as measured using qRT-PCR. B. The expression of let-7c and miR-99a was elevated in HepG2X and Hep3BX cells after knockdown of EZH2 by shRNAs. C. HepG2X and D Hep3BX cells handled with DZNep (10 µM) or DMSO (vehicle control) for 48 h. were executed to western blot analysis with anti-EZH2. The expression of let-7c and miR-99a was also determined in the same cells by qRT-PCR.
HBx-enhanced EZH2 expression and increases the binding in the let-7 promoter
miRNA expression has an effect on epigenetics [31] as well as histone and DNA methylation are coadjutant associated with epigenetic regulatory mechanisms [32]. For the purpose of exploring the underlying mechanism of let-7c dysregulation by EZH2, we first identified whether EZH2 can bind the let-7c promoter. Moreover, to evaluate the efficacy of upregulated EZH2 on the let-7c promoter, a chromatin immunoprecipitation (ChIP) assay was executed. Two separate primer pairs were constructed that are both specific for the let-7c promoter. The ChIP consequences illustrated that EZH2 was directly relative to the let-7c promoter (Fig. 5). These findings declared that HBx-upregulated EZH2 may strengthen the binding in the let-7 promoter.
HMGA2 is a target of let-7c
let-7c has been described to target HMGA2, a noticeable cellular oncogene [33]. Therefore, we investigated whether let-7c could downregulate HMGA2. We heightened let-7c expression through transfecting a let-7c mimic into HepG2X and Hep3BX cells while applying a mimic control as the negative control. A notable decline in the expression of HMGA2 protein was discovered after the upregulation of let-7c (Fig. 6A, B). Later, we examined whether the expression of HMGA2 corresponds to HBx expression in cells. HMGA2 mRNA levels were extraordinarily raised in HepG2X and Hep3BX cells in contrast to the parental cells (Fig. 6C). Likewise, HMGA2 protein levels were also significantly raised in HepG2X and Hep3BX cells (Fig. 6D). Furthermore, we adopted a plasmid encoding EZH2 into HepG2 and Hep3B cells. The result revealed that the level of HMGA2 was remarkably upraised in EZH2-expressing HepG2 and Hep3B cells in contrast to the empty vector (Fig. 6E).
HMGA2 is a target of let-7c. A, B. The comparative HMGA2 protein levels in HepG2X and Hep3BX cells 48 h after transfection with the let-7c mimic or the miRNA mimic control (negative control, NC). α-Tubulin was adopted as the endogenous control. C, D. Differential expression of HMGA2 mRNA and protein was characterized in HepG2, HepG2X, Hep3B, and Hep3BX cells. E EZH2 was transfected into HepG2 and Hep3B cells. HMGA2 expression, which was measured using qRT-PCR, was normalized against β-actin
HBx promotes the migration of HCC cells via HMGA2
To evaluate whether HBx mediates a heightening in the migration of HCC cells, we first compared the migration ability with Hep3B and Hep3BX cells. Our consequences confirmed that HBx accelerates cell migration (Fig. 7A). Moreover, the regulatory effect of HMGA2 on migratory capability was inspected with the transwell migration assay. Knockdown of HMGA2 with two individual shRNAs blocked the migration of Hep3BX cells (Fig. 7B). These data denoted that HMGA2 stands in the regulation of HCC cell migration.
Discussion
In recent years, there has been significant progress and improvement in the research and treatment of hepatocellular carcinoma (HCC). Single-cell approaches are powerful tools for studying HCC at a cellular level. They enable the investigation of intracellular signaling pathways in individual tumor cells, providing insights into cellular heterogeneity and signaling network activation. Shi et al. (2012) developed a single-cell proteomic chip that allowed the profiling of signaling pathways in HCC cells. This technology identified cell-to-cell variations in pathway activation, revealing the heterogeneity of HCC. Advancements in single-cell technologies, such as mass cytometry and single-cell RNA sequencing (scRNA-seq), have further expanded our understanding of HCC signaling networks [34]. Mass cytometry enables the measurement of multiple protein markers in individual cells, facilitating the analysis of signaling pathway activation across different HCC cell types [35]. scRNA-seq has been extensively used to study HCC, allowing for the simultaneous profiling of gene expression in thousands of cells and providing insights into cell populations and communication networks. It has been demonstrated that the application of scRNA-seq in analyzing the tumor microenvironment and drug efficacy in HCC, uncovering specific cellular interactions and molecular signatures [36]. Therefore, single-cell approaches enable the study of cellular heterogeneity, identification of rare cell populations, and characterization of signaling protein activation at a single-cell resolution, ultimately advancing our understanding of HCC development, progression, and therapeutic responses. Hepatitis B x antigen (HBx) has an extensive aspect of functions for regulating many genes in the host, it participates in the signaling pathways such as cell proliferation, cell cycle, metastasis, invasion, and protein degradation [37]. In our previous research, HBx was reported to be associated with HCC cell migration [10]. Therefore, HBx is a critical risk factor and a therapeutic target for the development of HCC [38]. In this study, we observed a significantly higher mRNA expression of EZH2 in most tumor samples in a contrast to non-tumor tissues. Similarly, Sasaki and Sudo also declared that EZH2 is highly expressed in cancer tissues and is closely related to the degree of tumor malignancy [39, 40]. Additionally, Xiao-Yan Shi et al. found that HBx raises the expression of EZH2 in hepatocytes through activation of the transcription factor E2F1 [41]. There are results that HBx can induce liver fibrosis also through the inhibition of the EZH2 complex [25]. There are results that HBx can induce liver fibrosis also through the inhibition of the EZH2 complex. And also, EZH2 complex acts as a pivotal regulator of epigenetics in HCC and promotes tumor growth and metastasis during HCC development [42, 43].
miRNA dysregulation can be discovered in many classes of cancer, and involved in the invasion and metastasis of cancer. Hence, miRNAs may act as tumor suppressors or oncogenes [44]. It is known that EZH2 as an epigenetic regulator, is involved in solid tumors progression and metastasis. EZH2 was reported to inhibit let-7c, miR-101, miR-125b, miR-139, and miR-200b [25, 45, 46]. In this study, we searched out the regulatory mechanism of EZH2 and discovered that HBx can depress the expression of cellular miRNAs (let-7c, miR-199a, and miR-99a) to adjust these critical cellular pathways. Henry et al. reported a remarkable reduction in miR-199a-3p expression and targeted by rapamycin (mTOR) and c-Met in HCC cell lines [47]. Hence, miR-199a-3p was shown to own tumor suppressor function. In addition, miR-99a has commonly been restrained in various tumors such as tongue squamous cell carcinoma, lung cancer, bladder, and prostate cancer [48,49,50,51]. And, the restrained capability of miR-99a on the development of HCC has been illustrated in vitro and in vivo [47]. Furthermore, IGF-1R and mTOR are first-hand objectives of miR-99a, indicating the property of miR-99a as a cell cycle inhibitor [52]. let-7, the first miRNA discovered in humans, involves in many cancer-related genes such as c-MYC, HMGA2, and CCND1 [33]. The functions of let-7 are well known to participate in the development, proliferation, and differentiation. According to our consequences, we found that HBx and EZH2 were highly expressed in HBV-infected HCC, and EZH2 adjusts methylation of the let-7 promoter, thereby suppressing the level of let-7.
HMGA2 binds to AT-rich regions in DNA and converts chromatin structure. It also interacts with certain proteins, such as E2 promoter-binding factor (E2F1) combing to create a complex to rise gene transcription [53]. During early developmental periods, HMGA2 is widely presented in many undifferentiated cells. AS fetal development goes on, the level of HMGA2 reduces [54,55,56]. Nevertheless, in most malignant tumors, the expression of HMGA2 raises, such as breast, lung, and pancreas cancer [57,58,59]. Importantly, the increased expression of HMGA2 is relative to accelerating angiogenesis, strengthening EMT, invasion, and metastasis in cancer progression [53, 60]. HMGA2 can effectively adjust EMT through the PI3K/AKT, MAPK/ERK, TGFB/Smad, NFrB, and STAT3 signaling pathways, enabling HCC cells to invade local tissues, get the penetrating ability of intravascular, and give birth to offspring with the tumor-initiating ability [53]. Therefore, in our results, the HMGA2 mRNA levels were obviously raised in HCC cells (HepG2X and Hep3BX) in contrast to the parental cells through the EZH2 - let-7c axis. We also discovered that the knockdown of HMGA2 restrained the migrated capability of Hep3BX. Besides, we confirmed that the down-regulation of let-7c causes the elevation of HMGA2 and improves the metastasis of HCC (Fig. 8). And, we provided new insights into the EZH2 epigenetic repression of let-7c that advances HCC migration caused by HBx. In summary, our research supported two markers, let-7c and HMGA2, as diagnostic markers and probable therapeutic objectives for HBV-infected HCC.
Conclusions
Our results provide new insights into the EZH2 epigenetic repression of let-7c that promotes HCC migration induced by HBx. Thus, let-7c and HMGA2 represent key diagnostic markers and potential therapeutic targets for the treatment of HBV-related HCC.
Methods
Patient characteristics
Serum samples were received from the National Cheng Kung University Hospital in Taiwan. Sample identified information was encrypted to guard patient confidentiality. And the samples obeyed the agreements approved by the Institutional Review Board of Cheng Kung University Hospital (assignment number: ER-99-176).
Comprehensive analysis of EZH2, let-7c, mir-199a-3p, and mir-99a from the Cancer Genome Atlas (TCGA)
Gene expression profile interactive analysis (GEPIA, http://gepia.cancer-pku.cn/index.html) uses standard processing pipelines to analyze RNA-Seq expression data from GTEx and TCGA. The Kaplan-Meier plotter can assess the survival rate of the effect of 54k genes (mRNA, miRNA, protein) and 21 cancer types (http://kmplot.com/analysis/). Sources for the databases include GEO, EGA, and TCGA. In this study, we used GEPIA for tumor/normal differential expression and overall survival analysis of EZH2 and the KM plotter for overall survival analysis of let-7c, mir-199a-3p, and mir-99a in HCC.
Microarray analysis
The mirVana miRNA Bioarray V9.2 was acquired from Ambion. The experimental method is according to the manufacturer’s protocol. Briefly, miRNAs were elicited with the mirVana™ miRNA Isolation Kit and were condensed with a flashPAGE™ Fractionator. After, small RNAs containing mature miRNAs were tagged with the mirVana™ miRNA Labeling Kit and hybridized to the mirVana™ miRNA Bioarray V9.2.
Cell Culture
HepG2 and Hep3B cell lines were gained from the American Type Culture Collection (Maryland), cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium supplemented with 10% fetal bovine serum (HyClone, Logan, UT). HepG2X and Hep3BX cells are apart derivatives of human hepatoma HepG2 and Hep3B cells, and stably express the gene encoding HBx, as reported previously [61].
Plasmids and miRNA mimic
The pcDNA-EZH2 plasmid was purched from Addgene (Cambridge, MA). The let-7c mimic and the miRNA mimic control were acquired from Dharmacon (Lafayette, CO). The miRNA mimic is a double-stranded oligonucleotide devised to mimic the function of an endogenous mature miRNA.
Transient transfection
Transfections were executed with Lipofectamine 2000, based on the manufacturer’s manual. After 48 h., cells were applied for subsequent related experiment [62].
Gene knockdown with shRNAs
Knockdown of genes was executed with specific shRNAs conveyed with the lentiviral system from the National RNAi Core Facility (Academia Sinica, Taipei, Taiwan) on the basis of their instruction manual. The shRNA constructs targeting HMGA2 are clones TRCN0000021966 and TRCN0000342671, and the construct targeting EZH2 is clone TRC0000040076. The shRNA construct against luciferase (shLuc), clone TRCN0000072244, was adopted as a negative control. The capability and the specificity were examined according to the previous report [11].
Quantitative PCR (qPCR)
Total RNA was extracted with TRIzol reagent (Invitrogen) based on the manufacturer’s instructions. The primers (Table 1) were synthesized by Invitrogen. qPCR was executed as previously described [11, 63], and detected with the LightCycler 480 apparatus (Roche).
Chromatin immunoprecipitation (ChIP) assay
A ChIP assay was executed by the MAGnify Chromatin Immunoprecipitation kit (Invitrogen). The experimental procedure was obeyed by the manufacturer’s manual. Finally, the break of chromatin was immunoprecipitated with EZH2 antibodies overnight at 4 °C. Two sets of primers were designed on the report of the let-7c promoter, and PCR acted as described [62]. The two primer pairs were as follows: primer pair 1, AGCCTTCTGAGCCAGTTTCTTC (forward) and CCCGAGCAGTAGCAGTGTG (reverse); primer pair 2, CGGGTGCTTTCTATCTCTTCTCC (forward) and TGGATGCCGTGGCTTCTCG (reverse).
Western blot analysis
The antibodies and chemicals were applied: anti-hepatitis B Virus X antigen (Abcam, Burlingame, CA, USA; 1:1000); anti-EZH2, clone AC22 (Cell Signaling; 1:1000); anti-HMGA2 (Abcam; 1:1000); and anti-β-actin (Sigma; 1:5000); Chemiluminescence detection reagent (GE, Piscataway, NJ). The experimental procedure refers to the previous report [11].
Transwell Migration assays
Cell migration was evaluated with Boyden chambers (Millipore, Billerica, MA). In brief, 1 × 105 cells in DMEM plus 1% FBS were seeded onto each membrane insert (8 μm). After 24 h incubation, the cells in the upper chamber were dismissed with a cotton swab. The invasive cells were then stained with Giemsa and calculated under a light microscope at 100x magnification [64, 65].
Statistical analysis
Detailed examination of data was calculated with Prism 8 (GraphPad) and Excel (Microsoft). Statistical analyses were carried out with a two-tailed Student’s t-test or Fisher’s exact test.
Data Availability
Not applicable.
References
Huang TY, Peng SF, Huang YP, Tsai CH, Tsai FJ, Huang CY, et al. Combinational treatment of all-trans retinoic acid (ATRA) and bisdemethoxycurcumin (BDMC)-induced apoptosis in liver cancer Hep3B cells. J Food Biochem. 2020;44:e13122.
Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–14.
McGlynn KA, Petrick JL, El-Serag HB. Epidemiol Hepatocellular Carcinoma Hepatol. 2021;73(Suppl 1):4–13.
Bayram AA, Al-Dahmoshi HOM, Al-Khafaji NSK, Al Mammori RTO, Al-Shimmery AHS, Saki M. Study of the D-dimer, C-reactive protein, and autoantibodies markers among HBV infected patients in Babylon province. Iraq Biomed (Taipei). 2021;11:67–72.
Su Q, Schroder CH, Hofmann WJ, Otto G, Pichlmayr R, Bannasch P. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology. 1998;27:1109–20.
Raita Y, Perez-Losada M, Freishtat RJ, Harmon B, Mansbach JM, Piedra PA, et al. Integrated omics endotyping of infants with respiratory syncytial virus bronchiolitis and risk of childhood asthma. Nat Commun. 2021;12:3601.
Shen Z, Wu J, Gao Z, Wang J, Zhu H, Mao R, et al. Characterization of IL-21-expressing recombinant hepatitis B virus (HBV) as a therapeutic agent targeting persisting HBV infection. Theranostics. 2020;10:5600–12.
Lara-Pezzi E, Majano PL, Yanez-Mo M, Gomez-Gonzalo M, Carretero M, Moreno-Otero R, et al. Effect of the hepatitis B virus HBx protein on integrin-mediated adhesion to and migration on extracellular matrix. J Hepatol. 2001;34:409–15.
Chung TW, Lee YC, Kim CH. Hepatitis B viral HBx induces matrix metalloproteinase-9 gene expression through activation of ERK and PI-3K/AKT pathways: involvement of invasive potential. FASEB J. 2004;18:1123–5.
Su KJ, Yu YL. Downregulation of SHIP2 by Hepatitis B Virus X promotes the Metastasis and Chemoresistance of Hepatocellular Carcinoma through SKP2. Cancers (Basel). 2019;11.
Wu CS, Yen CJ, Chou RH, Chen JN, Huang WC, Wu CY, et al. Downregulation of microRNA-15b by hepatitis B virus X enhances hepatocellular carcinoma proliferation via fucosyltransferase 2-induced Globo H expression. Int J Cancer. 2014;134:1638–47.
Mani SKK, Yan B, Cui Z, Sun J, Utturkar S, Foca A, et al. Restoration of RNA helicase DDX5 suppresses hepatitis B virus (HBV) biosynthesis and wnt signaling in HBV-related hepatocellular carcinoma. Theranostics. 2020;10:10957–72.
Feng J, Yang G, Liu Y, Gao Y, Zhao M, Bu Y, et al. LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics. 2019;9:5227–45.
Tang H, Oishi N, Kaneko S, Murakami S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006;97:977–83.
Lupberger J, Hildt E. Hepatitis B virus-induced oncogenesis. World J Gastroenterol. 2007;13:74–81.
Chou RH, Chiu L, Yu YL, Shyu WC. The potential roles of EZH2 in regenerative medicine. Cell Transpl. 2015;24:313–7.
Chou RH, Yu YL, Hung MC. The roles of EZH2 in cell lineage commitment. Am J Transl Res. 2011;3:243–50.
Hsieh YY, Lo HL, Yang PM. EZH2 inhibitors transcriptionally upregulate cytotoxic autophagy and cytoprotective unfolded protein response in human colorectal cancer cells. Am J Cancer Res. 2016;6:1661–80.
Su KJ, Lin CW, Chen MK, Yang SF, Yu YL. Effects of EZH2 promoter polymorphisms and methylation status on oral squamous cell carcinoma susceptibility and pathology. Am J Cancer Res. 2015;5:3475–84.
Yang PM, Hong YH, Hsu KC, Liu TP. p38alpha/S1P/SREBP2 activation by the SAM-competitive EZH2 inhibitor GSK343 limits its anticancer activity but creates a druggable vulnerability in hepatocellular carcinoma. Am J Cancer Res. 2019;9:2120–39.
Ruan Y, Xu H, Ji X, Zhao J. BLM interaction with EZH2 regulates MDM2 expression and is a poor prognostic biomarker for prostate cancer. Am J Cancer Res. 2021;11:1347–68.
Chang CJ, Hung MC. The role of EZH2 in tumour progression. Br J Cancer. 2012;106:243–7.
Shahhosseini A, Bourova-Flin E, Derakhshan S, Aminishakib P, Goudarzi A. High levels of histone H3 K27 acetylation and tri-methylation are associated with shorter survival in oral squamous cell carcinoma patients. Biomed (Taipei). 2023;13:22–38.
Zeng T, Luo L, Huang Y, Ye X, Lin J. Upregulation of miR-138 increases sensitivity to Cisplatin in Hepatocellular Carcinoma by regulating EZH2. Biomed Res Int. 2021;2021:6665918.
Au SL, Wong CC, Lee JM, Fan DN, Tsang FH, Ng IO, et al. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology. 2012;56:622–31.
Alzrigat M, Jernberg-Wiklund H. The miR-125a and miR-320c are potential tumor suppressor microRNAs epigenetically silenced by the polycomb repressive complex 2 in multiple myeloma. RNA Dis. 2017;4.
Bai B, Liu Y, Fu XM, Qin HY, Li GK, Wang HC, et al. Dysregulation of EZH2/miR-138-5p Axis contributes to Radiosensitivity in Hepatocellular Carcinoma Cell by Downregulating Hypoxia-Inducible factor 1 alpha (HIF-1alpha). Oxid Med Cell Longev. 2022;2022:7608712.
Yu YL, Chou RH, Shyu WC, Hsieh SC, Wu CS, Chiang SY, et al. Smurf2-mediated degradation of EZH2 enhances neuron differentiation and improves functional recovery after ischaemic stroke. EMBO Mol Med. 2013;5:531–47.
Yu YL, Chou RH, Chen LT, Shyu WC, Hsieh SC, Wu CS, et al. EZH2 regulates neuronal differentiation of mesenchymal stem cells through PIP5K1C-dependent calcium signaling. J Biol Chem. 2011;286:9657–67.
Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–63.
Iorio MV, Piovan C, Croce CM. Interplay between microRNAs and the epigenetic machinery: an intricate network. Biochim Biophys Acta. 2010;1799:694–701.
Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–4.
Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–30.
Shi Q, Qin L, Wei W, Geng F, Fan R, Shin YS, et al. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc Natl Acad Sci U S A. 2012;109:419–24.
Lun XK, Bodenmiller B. Profiling cell signaling networks at single-cell resolution. Mol Cell Proteomics. 2020;19:744–56.
Mi H, Ho WJ, Yarchoan M, Popel AS. Multi-scale spatial analysis of the Tumor Microenvironment reveals features of Cabozantinib and Nivolumab Efficacy in Hepatocellular Carcinoma. Front Immunol. 2022;13:892250.
Zhang WY, Cai N, Ye LH, Zhang XD. Transformation of human liver L-O2 cells mediated by stable HBx transfection. Acta Pharmacol Sin. 2009;30:1153–61.
Medhat A, Arzumanyan A, Feitelson MA. Hepatitis B x antigen (HBx) is an important therapeutic target in the pathogenesis of hepatocellular carcinoma. Oncotarget. 2021;12:2421–33.
Sasaki M, Ikeda H, Itatsu K, Yamaguchi J, Sawada S, Minato H, et al. The overexpression of polycomb group proteins Bmi1 and EZH2 is associated with the progression and aggressive biological behavior of hepatocellular carcinoma. Lab Invest. 2008;88:873–82.
Sudo T, Utsunomiya T, Mimori K, Nagahara H, Ogawa K, Inoue H, et al. Clinicopathological significance of EZH2 mRNA expression in patients with hepatocellular carcinoma. Br J Cancer. 2005;92:1754–8.
Shi XY, Zhang YY, Zhou XW, Lu JS, Guo ZK, Huang PT. Hepatitis B virus X protein regulates the mEZH2 promoter via the E2F1-binding site in AML12 cells. Chin J Cancer. 2011;30:273–9.
Feng GX, Li J, Yang Z, Zhang SQ, Liu YX, Zhang WY, et al. Hepatitis B virus X protein promotes the development of liver fibrosis and hepatoma through downregulation of miR-30e targeting P4HA2 mRNA. Oncogene. 2017;36:6895–905.
Yamaguchi H, Hung MC. Regulation and role of EZH2 in Cancer. Cancer Res Treat. 2014;46:209–22.
Zhang PP, Wang XL, Zhao W, Qi B, Yang Q, Wan HY, et al. DNA methylation-mediated repression of miR-941 enhances lysine (K)-specific demethylase 6B expression in hepatoma cells. J Biol Chem. 2014;289:24724–35.
Chen Z, Xiang L, Hu Z, Ou H, Liu X, Yu L, et al. Epigenetically silenced linc00261 contributes to the metastasis of hepatocellular carcinoma via inducing the deficiency of FOXA2 transcription. Am J Cancer Res. 2021;11:277–96.
Xu X, Gu J, Ding X, Ge G, Zang X, Ji R, et al. LINC00978 promotes the progression of hepatocellular carcinoma by regulating EZH2-mediated silencing of p21 and E-cadherin expression. Cell Death Dis. 2019;10:752.
Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, et al. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184–93.
Cheng H, Xue J, Yang S, Chen Y, Wang Y, Zhu Y, et al. Co-targeting of IGF1R/mTOR pathway by miR-497 and miR-99a impairs hepatocellular carcinoma development. Oncotarget. 2017;8:47984–97.
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of Tongue. Clin Cancer Res. 2008;14:2588–92.
Yamada H, Yanagisawa K, Tokumaru S, Taguchi A, Nimura Y, Osada H, et al. Detailed characterization of a homozygously deleted region corresponding to a candidate tumor suppressor locus at 21q11-21 in human lung cancer. Genes Chromosomes Cancer. 2008;47:810–8.
Catto JW, Miah S, Owen HC, Bryant H, Myers K, Dudziec E, et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009;69:8472–81.
Li D, Liu X, Lin L, Hou J, Li N, Wang C, et al. MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J Biol Chem. 2011;286:36677–85.
Mansoori B, Mohammadi A, Ditzel HJ, Duijf PHG, Khaze V, Gjerstorff MF et al. HMGA2 as a critical Regulator in Cancer Development. Genes (Basel). 2021;12.
Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910.
Monzen K, Ito Y, Naito AT, Kasai H, Hiroi Y, Hayashi D, et al. A crucial role of a high mobility group protein HMGA2 in cardiogenesis. Nat Cell Biol. 2008;10:567–74.
Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell. 2008;135:227–39.
Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11:R7.
Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–11.
Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating akt. Cancer Cell. 2009;16:259–66.
Cui H, Song R, Wu J, Wang W, Chen X, Yin J. MicroRNA-337 regulates the PI3K/AKT and Wnt/beta-catenin signaling pathways to inhibit hepatocellular carcinoma progression by targeting high-mobility group AT-hook 2. Am J Cancer Res. 2018;8:405–21.
Yen CJ, Lin YJ, Yen CS, Tsai HW, Tsai TF, Chang KY, et al. Hepatitis B virus X protein upregulates mTOR signaling through IKKbeta to increase cell proliferation and VEGF production in hepatocellular carcinoma. PLoS ONE. 2012;7:e41931.
Chien YC, Liu LC, Ye HY, Wu JY, Yu YL. EZH2 promotes migration and invasion of triple-negative breast cancer cells via regulating TIMP2-MMP-2/-9 pathway. Am J Cancer Res. 2018;8:422–34.
Achudhan DCSL, Liu SC, Lin YY, Huang WC, Wu YC, Huang CC, Tsai CH, Ko CY, Kuo YH, Tang CH. Antcin K inhibits VCAM-1-dependent monocyte adhesion in human rheumatoid arthritis synovial fibroblasts. Food & Nutrition Research. 2022;66:8645.
Chang WS, Tsai CW, Yang JS, Hsu YM, Shih LC, Chiu HY, et al. Resveratrol inhibited the metastatic behaviors of cisplatin-resistant human oral cancer cells via phosphorylation of ERK/p-38 and suppression of MMP-2/9. J Food Biochem. 2021;45:e13666.
Lee KT, Su CH, Liu SC, Chen BC, Chang JW, Tsai CH et al. Cordycerebroside A inhibits ICAM-1-dependent M1 monocyte adhesion to osteoarthritis synovial fibroblasts. J Food Biochem. 2022:e14108.
Acknowledgements
Not applicable.
Funding
This research was funded by grants from the National Science and Technology Council, Taiwan (NSTC 109-2320-B-039-013-MY3; NSTC 110-2314-B-039-048 -; NSTC 111-2314-B-039-070-; NSTC 110-2314-B-039 -034 -MY3; NSTC 112-2320-B-039-020), the National Health Research Institute, Taiwan (NHRI-112A1-CACO-13232302), the China Medical University, Taiwan (CMU108-MF-01; CMU109-MF-03), the China Medical University Hospital, Taiwan (DMR-111-053; DMR-111-209; DMR-112-198; DMR-112-195).
Author information
Authors and Affiliations
Contributions
C.S.W., Y.C.C., and C.J.Y.; formal analysis, J.Y.W., L.Y.B., C.J.Y., and Y.L.Y.; resources and software, Y.C.C., C.S.W., C.J.Y., and J.Y.W.; writing—original draft and methodology, C.S.W., Y.C.C., and Y.L.Y.; supervision and writing—review and editing, Y.L.Y., Y.C.C., and L.Y.B. All authors read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of Cheng Kung University Hospital (assignment number: ER-99-176).
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wu, CS., Chien, YC., Yen, C. et al. EZH2-mediated epigenetic silencing of tumor-suppressive let-7c/miR-99a cluster by hepatitis B virus X antigen enhances hepatocellular carcinoma progression and metastasis. Cancer Cell Int 23, 199 (2023). https://doi.org/10.1186/s12935-023-03002-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-023-03002-9