Abstract
Colorectal cancer (CRC) is the 3rd most common type of cancer worldwide. Late detection plays role in one-third of annual mortality due to CRC. Therefore, it is essential to find a precise and optimal diagnostic and prognostic biomarker for the identification and treatment of colorectal tumorigenesis. Covalently closed, circular RNAs (circRNAs) are a class of non-coding RNAs, which can have the same function as microRNA (miRNA) sponges, as regulators of splicing and transcription, and as interactors with RNA-binding proteins (RBPs). Therefore, circRNAs have been investigated as specific targets for diagnostic and prognostic detection of CRC. These non-coding RNAs are also linked to metastasis, proliferation, differentiation, migration, angiogenesis, apoptosis, and drug resistance, illustrating the importance of understanding their involvement in the molecular mechanisms of development and progression of CRC. In this review, we present a detailed summary of recent findings relating to the dysregulation of circRNAs and their potential role in CRC.
Similar content being viewed by others
Background
Colorectal cancer (CRC) is one of the most common malignancies ranking third in the incidence and second in mortality among other cancers in the world. The global incidence of CRC is increasing, with approximately 3640 deaths and 17,930 new cases in 2020 [1, 2]. The exact mechanisms underlying CRC development remain unknown, however, risk factors that are strongly related to CRC include genetics, diet, tobacco smoking, heavy alcohol consumption, inactive lifestyle and age, where > 50 is a significant risk factor for CRC. However, recent evidence has also detected an increased risk for young adults [3]. Clearly the disorder is multifactorial in nature, with no common identifiable predictor of pre-disposition [4]. Here, we will review the molecular evidence to date.
Genetic and epigenetic alterations have both been found in CRC patients; changes in chromosomal copy number, aberrant gene methylation, and dysregulated gene expression, including tumor suppressor genes such as APC, BRAF, DCC, TP53, SMAD4, SMAD2, oncogenes such as KRAS and NRAS, and DNA repair genes including MLH1 and MSH6 [5, 6].
Dividing these mutation types into functional pathways broadly identifies three separate mechanisms: Chromosomal instability, which is the most common cause of genomic instability in CRC, significantly linked to alterations in APC and KRAS genes [7, 8]. In hereditary and sporadic colorectal cancer, microsatellite instability (MSI) is another key pathway. Germline mutation in one of the DNA mismatch repair genes, MLH1, MSH2, MSH6, or PMS2 leads to hereditary nonpolyposis colorectal cancer (HNPCC), while MSI in sporadic colorectal cancer is predominantly due to hypermethylation of the MLH1 promoter and sometimes sporadic mutations [9]. Defects in the mismatch repair mechanisms can also lead to MSI status [10]. A third pathway is via epigenetic alteration. CpG island methylator phenotype (CIMP) differences can result in changes in gene expression or function without changing the DNA sequence of that particular gene [11]. Taken together; these three pathways indicate the genetic heterogeneity of CRC.
CRCs are classified into 4 subtypes: CMS1-CMS4 with different clinical and biological characterizations [12]. Despite recent advances in our knowledge of signaling pathways involved in CRC, chemo- and radiotherapy resistance remains the most significant hurdle in CRC treatment. Therefore, a novel methodology for improved early diagnosis is essential. Non-coding RNAs (ncRNAs) play important roles in the regulation of chemo-and radio resistance of CRC [13]. Thus, ncRNAs could serve as targets for the development of new therapeutic strategies for drug and radiation resistance in CRC [14, 15]. circRNAs are a significant facet in ncRNAs biology, thus understanding of the role of circRNAs in CRC progression is pivotal to identifying new diagnostic, prognostic and predictive biomarkers for CRC [16]. In this review, we summarize the potential clinical implications of human circRNAs in CRC, for use as predictive biomarkers and/or therapeutic targets.
The non-coding RNAs
The majority of the human genome (~ 90%) is transcribed as ncRNAs, which contain multiple classes of RNAs with various lengths [17]. Many studies have identified functional roles for ncRNAs, in various physiological and pathological processes, such as diabetes, cardiovascular disease, and cancer [18,19,20]. Classes of short ncRNAs include microRNAs (miRNAs), small interfering RNAs (siRNAs) and short piwi-interacting RNAs (piRNAs), meanwhile, linear lncRNAs (long non-coding RNAs) and circular RNAs are both classed as long noncoding RNAs [21]. circRNAs, however, are a new class of long ncRNAs, processing largely from exotic or intronic sequences, and are remarkably unique in structure and chemical characteristics compared with linear RNAs. circRNA biogenesis is based on the back‐splicing process, and closed 5-3ʹ ends negate degradation by RNA exonuclease or RNase R [22]. Classification of circRNAs is largely based on sequence origin, where subgroups include the circular intronic RNAs (ciRNAs), the exonic circRNAs (EcircRNAs), and exon–intron circRNAs (EIciRNAs) [23]. EcircRNAs, which predominantly exist in the cytoplasm, comprise the majority of all circRNAs. EcircRNAs can be formed by three different mechanisms, including lariat-driven circularization, RNA-binding protein (RBP)-driven circularization, and back splicing. EIciRNAs however, are formed only by back splicing of ciRNAs, which depends on a 7-nt GU-rich element and an 11-nt C-rich element, important in escaping debranching and exonucleolytic degradation [23, 24]. circRNAs have relatively stable structure and show tissue-specific expression, also displaying developmental stage regulation, with evolutionary conservation among species [25].
Functions of circRNAs
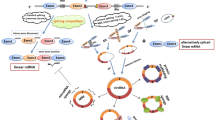
circRNAs have regulatory roles in gene expression by sponging miRNAs, competing with other RNAs for binding to miRNAs and RNA binding proteins (RBPs) to modulate the local concentration of RBPs and RNAs as part of the competing endogenous RNA (ceRNA) network [26]. circRNAcircRNACDR1as (ciRS-7), for example, which has more than 70 conserved binding sites for miR-7, and is highly expressed in human and mouse brains [27, 28]. SRY, which encodes both linear and circular RNAs, is involved in sex determination in testis development. circRNA SRY can control metastasis and invasion of tumor cells via sponging miR-138 [29, 30]. Another circRNA, known as CircITCH, plays similar roles as a miRNA sponge, via miR-7, miR-17, and miR214, to inhibit proliferation through the Wnt/β-catenin signaling pathway [31], which is illustrated in Fig. 1A.
(A) Circ0001313 was found to be the most significantly upregulated circular RNA in CRC. where it can sponge miR-338 and affect apoptosis radiosensitivity in CRC. (B) Circ-ITCH is overexpressed in colorectal cancer and it can develop proliferation by sponging miR7, miR214, miR17 via Wnt/ β-catenin pathway signaling
Although circRNAs are considered to be non-coding RNAs due to lack of 5’-cap structure and 3’-polyadenylation tail, circRNAs have been shown to generate protein products in a cap-independent manner [32]. Interestingly, many circRNAs are sometimes translated, indeed using high-content genomic screening, Legnini et al. found Circ-ZNF609 can translate into a protein in a splicing-dependent and cap-independent manner [33]. Yang Y et al. discovered CircFBXW7, produced from the FBXW7 gene, encoding a novel 21-kDa protein FBXW7-185aa, which reduced the half-life of c-Myc by antagonizing USP28-induced c-Myc stabilization [34].
The overall activities of circRNAs are intricately intertwined with RNA binding proteins, modulating the stability of mRNAs, regulating gene transcription, and translating proteins [35] and are involved in the regulation of cell proliferation, pluripotency and early lineage differentiation, epithelial-mesenchymal transition (EMT), cancer progression and chemoradiotherapy resistance, as shown in Fig. 2.
Upregulation of circRNAs in CRC
Among all the validated aberrantly expressed circRNAs in colorectal cancer, upregulation of circRNAs more often associates with oncogenesis. Xia et al. found abnormally expressed circRNAs through CircRNA high-throughput sequencing, identifying Circ-0053277 as having the ability to sponge miR‐2467‐3p, and as being significantly upregulated in CRC tissues, where it facilitated CRC cell migration, proliferation, and epithelial‐mesenchymal transition [36]. Similarly, Li et al. identified CircVAPA as being upregulated in tissues and plasma, serving as a sponge for miR-101. Furthermore, they showed that the expression level of miR-125a was decreased in CRC cells, and CircVAPA knockdown repressed CRC cells cycle progression, invasion, and migration [37]. Knockdown of CircVAPA can also suppress CRC cell cycle progression, invasion, and migration by sponging miR-125a [38].
Yahang et al. found that Hsa_Circ_0026416 which was upregulated in CRC tissues and plasma, and has a key role in promoting the progression of CRC both in vitro and in vivo, may function as a ceRNA to sponge miR-346 [39].
Knockdown of another upregulated circRNA, CircACAP2 (hsa_circ_0007331), which was reported to be significantly upregulated in CRC tissues and colon cancer cells lines, suppressed proliferation and invasion by downregulating T lymphoma invasion and metastasis protein 1 (Tiam1) expression, through upregulated miR-21-5p expression (40). Another highly overexpressed circRNA in CRC is Hsa_circ_0136666, derived from the PRKDC gene, which can regulate proliferation and migration of CRC cells by sponging miR-136 [41].
Downregulated circRNAs in CRC
As well as being overexpressed, other circRNAs are downregulated in CRC. Wang X et al. showed hsa_Circ_001988 was significantly downregulated in 31 matched colorectal cancer tissue samples, proposing this circRNA as a novel diagnosis potential biomarker in the CRC [42]. Geng Y reported hsa_Circ_0009361 to be significantly downregulated in both CRC tissues and derived cells. circRNA promoting the proliferation, epithelial-mesenchymal transition, migration, and invasion of CRC cells by sponging of miR-582. Conversely, overexpression of hsa_Circ_0009361 caused upregulation in the expression of adenomatous polyposis coli 2 (APC2) and blocked the activity of the Wnt/β-catenin pathway [43]. Circ-ITGA7, which sponges’ miR-370-3p to increase ITGA7 transcription–, through inhibition of RREB1 via oncogenic Ras has been shown to be down-regulated in CRC tissue samples [44]. Indeed, Circ-ITGA7 has also been shown to directly act as a tumor suppressor in CRC, with clinical features including cancer differentiation, lymph node metastasis, distant metastasis, and alterations in the TNM stage [45]. circRNA Circ-FBXW7 silencing was previously reported to enhance the proliferation, cell migration, and invasion of CRC cells in culture. In contrast, overexpression of Circ-FBXW7 significantly suppressed CRC cell proliferation, migration, and invasion. Similarly, Circ-FBXW7 silencing was also shown to stimulate tumor growth in SW480 and SW620 tumor models, whereas Circ-FBXW7 overexpression repressed tumor progression in the same system. This suggests that Circ-FBXW7 could serve as a target biomarker of CRC. Potential mechanisms have been proposed, including upregulated mRNA and protein expressions of NEK2 and mTOR, and diminished the PTEN expression (46). circRNACirc_021977 is another circRNA found to be down-regulated in CRC. Circ_021977 was shown to sponge miR-10b-5p, with a regulatory axis inhibiting proliferation, migration, and invasion in CRC via p21 and p53 [47]. Dysregulated circRNA expression in CRC is summarized in Table 1.
circRNAs in predicting response to chemoradiotherapy
Targeted therapy, chemotherapy, and multiagent regimens, for example, FOLFIRI (5-FU and irinotecan) and FOLFOX (5-FU oxaliplatin) can be applied as the standard treatment of CRC. However, chemotherapy has its limitations, including toxicity, low response rates, unpredictable innate and acquired resistance mechanisms, and low tumor-specific selectivity [137]. Recent studies have shown that different ncRNAs such as circRNAs, may play important roles in the regulation of chemoresistance and affect the sensitivity of tumors to chemotherapy and radiotherapy through modification of various signaling pathways, including cell cycle, proliferation, apoptosis, and DNA damage repair [84, 112]. hsa_circRNA_0001313 is one of the upregulated circRNAs in radio-resistant CRC tissues. Inhibition of hsa_circRNA_0001313 induces radio-sensitivity, reduced cell viability, and increases caspase-3 activity and colony formation by negatively modifying miR-338-3p in CRC cells, which has shown in Fig. 1B [124]. Another recent study reported that CircDDX17 was down-regulated in CRC, and its overexpression induced inhibition of 5-Fu resistance, blocked tumor growth, and CRC progression via sponging miR-31-5p [131]. Interestingly, Circ-32883 was upregulated in CRC tissues and its overexpression was positively associated with chemoresistance through its potential action as a sponge for miR-501-5p. This miRNA binds to EML5 mRNA, inhibiting its expression. Thus, promoting resistance to FOLFOX therapy [112]. Other circRNAs related to chemotherapy resistance are summarized in Table 2.
circRNAs as biomarkers for colorectal cancer
Through improvements in high-throughput sequencing, circRNA microarray, and chip analysis we now know circRNAs are differentially expressed in CRC, and certain circRNAs are involved in various biological processes such as proliferation, migration, invasion, and apoptosis. Due to the unique structure of circRNAs, which confers resistance to RNase and longer half-lives, they can therefore be potential candidates for diagnostic biomarkers. However, the underlying biological function of circRNAs requires further investigation [138, 139].
Several circRNAs have been proposed as useful therapeutic targets for CRC. For instance, hsa_circ_022382 which is derived from the human FADS2 gene is overexpressed in 200 CRC tissues, where CircFADS2 overexpression was positively associated with clinicopathological features. CircFADS2 expression may therefore be a promising biomarker for prognostic investigation in CRC patients [95]. In another study, hsa_circ_0026344 was shown to be significantly down-regulated in 32 CRC patients compared to paired adjacent non-tumorous tissues. The expression of hsa_circ_0026344 was correlated with tumor size and lymph metastasis. Functionally, circRNA-0026344 overexpression significantly suppressed CRC cell proliferation and colony formation as well as promoted apoptosis by regulating miR-21 and miR-31 levels [45]. Other circRNAs with biomarker potential are summarized in Table 3.
circRNAs as therapeutic targets in colorectal cancer
Targeted therapy has been widely used in the clinic due to its excellent efficacy, and it can work on cancerous cells by directly inhibiting cell proliferation, differentiation, and migration [50]. Indeed, monoclonal antibodies, for instance, are currently an important player in targeted therapies [51]. circRNAs moderate drug resistance by sponging microRNAs both in traditional chemotherapeutic drugs, advanced targeted drugs, and immunotherapeutic drugs. For example, therapeutic targeting of ciRS-7 may become a promising strategy for colorectal cancer patients, since higher expression of ciRS-7 correlated with multiple clinicopathologic factors, such as advanced T-stage, lymph node, and distant metastasis, and ciRS-7 overexpression promotes the EGFR/RAF1/MAPK pathway by inhibiting miR-7 activity [121, 155]. Yang et al. indicated that high expression of circPTK2 positively correlated with poorer survival, showing CircPTK2 can bind to vimentin and promote EMT growth and metastasis in CRC cells, therefore ciRS-7 may become a therapeutic target for CRC metastasis [51]. The relation between circPTK2 in CRC is shown in Fig. 3.
Another highly expressed circRNA in CRC tissue is Circ_001680 which was observed to enhance the proliferation and migration capacity of CRC cells. Fluorescence reporter assays confirmed that circ_001680 alters the expression of BMI1 by targeting miR-340. More importantly, Circ_001680 was found to promote the propogation of cancer stem cells in CRC and induce resistance against Irinote by modifying the miR-340 target gene BMI1 n [53]. Safe and effective delivery of ncRNAs is a significant therapeutic paradigm for all cancers. Since unmodified oligonucleotides are not stable in circulation, modifications of oligonucleotides are essential to increasing efficacy and stability. Most current oligonucleotide therapies need an additional delivery system to achieve these desired biological effects. Several options need to be considered in selecting a delivery system, including stability, evasion of the innate immune system, avoidance of non-specific interactions with serum proteins, and non-target cells. One of the common strategies to increase the circulation time for therapeutic oligonucleotides is shielding the exterior of delivery vehicles with polyethylene glycol (PEG). This strategy may prevent the non-specific function of particles with immune cells and other non-target tissues. Although a variety of delivery systems has been developed in the laboratory, challenges remain in bringing the full potential of RNAi to clinical approaches [156]. circRNAs however, offer significant increases in stability over current strategies.
Conclusions and perspectives
Following advancements in high-throughput sequencing, the field of circRNAs has attracted more attention and is currently an area of intense interest in the field of cancer research. circRNAs are an ideal biomarker in cancer, and are stably expressed in exosomes, blood, and saliva, where specific circRNAs have been indicated as promising prognostic or diagnostic biomarkers already.
Abnormal expression of circRNAs has been observed in a wide range of human malignancies and their dysregulation can alter gene expression networks, leading to dramatic changes in cell fates, including cancer initiation and progression. circRNAs can be both oncogenic and anti-oncogenic, so could potentially be utilized in the treatment and prognosis of colorectal cancer. Although recent advances on circRNAs have highlighted some interesting insights, much work remains to be done to translate circRNAs into clinical application for clinical patient benefit. Major hurdles include the development of an efficient siRNAs delivery system, and the assessment of safety and side effects, yet, clearly circRNAs have significant potential for the treatment and diagnosis of CRC.
Availability of data and materials
Data will be provided based on reasonable request.
Abbreviations
- ceRNA:
-
Competing endogenous RNA
- circRNAs:
-
Circular RNAs
- siRNA:
-
Small interacting RNA
- ncRNA:
-
Noncoding RNA
- HNPCC:
-
Hereditary nonpolyposis colorectal cancer
- ciRNAs:
-
Intronic circRNAs
- ecircRNA:
-
Exonic circRNAs
- ELciRNA:
-
Exon–intron-circRNAs
- miRNA:
-
MicroRNA
- RBP:
-
RNA-binding protein
- PEG:
-
Polyethylene glycol
- CRC:
-
Colorectal cancer
References
Keum NN, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16(12):713–32.
Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–64.
Zheng Y, Hua X, Win AK, MacInnis RJ, Gallinger S, Le Marchand L, et al. A new comprehensive colorectal cancer risk prediction model incorporating family history, personal characteristics, and environmental factors. Cancer Epidemiol Biomarkers Prev. 2020;29(3):549–57.
Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, et al. Colorectal cancer. Nat Rev Dis Prim [Internet]. 2015;1:15065.
Koveitypour Z, Panahi F, Vakilian M, Peymani M, Seyed Forootan F, Nasr Esfahani MH, et al. Signaling pathways involved in colorectal cancer progression. In: Cell and bioscience, vol. 9. BioMed Central Ltd.; 2019. https://pubmed.ncbi.nlm.nih.gov/31827763/
Gausachs M, Borras E, Chang K, Gonzalez S, Azuara D, Amador AD, et al. Mutational heterogeneity in APC and KRAS arises at the crypt level and leads to polyclonality in early colorectal tumorigenesis. Clin Cancer Res. 2017;23(19):5936–47.
APC, K-ras, and p53 gene mutations in colorectal cancer patients: correlation to clinicopathologic features and postoperative surveillance—PubMed. https://pubmed.ncbi.nlm.nih.gov/15943410/
Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138(6):2059–72.
Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18(1):197. https://doi.org/10.3390/ijms18010197.
Collura A, Lefevre JH, Svrcek M, Tougeron D, Zaanan A, Duval A. Microsatellite instability and cancer: from genomic instability to personalized medicine. Medecine/sciences, vol. 35. Editions EDK; 2019. p. 535–43. https://pubmed.ncbi.nlm.nih.gov/31274083/
Wang X, Wang D, Zhang H, Feng M, Wu X. Genome-wide analysis of DNA methylation identifies two CpG sites for the early screening of colorectal cancer. Epigenomics. 2020;12(1):37–52.
Okita A, Takahashi S, Ouchi K, Inoue M, Watanabe M, Endo M, et al. Consensus molecular subtypes classification of colorectal cancer as a predictive factor for chemotherapeutic efficacy against metastatic colorectal cancer. Oncotarget. 2018;9(27):18698–711.
Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochemical journal, vol. 474. Portland Press Ltd; 2017. p. 4219–51. https://pubmed.ncbi.nlm.nih.gov/29242381/
Matsui M, Corey DR. Non-coding RNAs as drug targets. Nature reviews drug discovery, vol. 16. Nature Publishing Group; 2017. p. 167–79. https://pubmed.ncbi.nlm.nih.gov/27444227/
Panni S, Lovering RC, Porras P, Orchard S. Non-coding RNA regulatory networks. Biochimica et Biophysica Acta—gene regulatory mechanisms, vol. 1863. Elsevier B.V.; 2020. https://pubmed.ncbi.nlm.nih.gov/31493559/
Hao S, Cong L, Qu R, Liu R, Zhang G, Li Y. Emerging roles of circular RNAs in colorectal cancer. OncoTargets and therapy, vol. 12. Dove Medical Press Ltd.; 2019. p. 4765–77. https://pubmed.ncbi.nlm.nih.gov/31354303/
Akhade VS, Pal D, Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Adv Exp Med Biol. 2017;1008:47–74.
Ameli-Mojarad M, Ameli-Mojarad M, Nourbakhsh M, Nazemalhosseini-Mojarad E. Circular RNA hsa_circ_0005046 and hsa_circ_0001791 may become diagnostic biomarkers for breast cancer early detection. J Oncol. 2021;2021:1–7.
Tian D, Xiang Y, Tang Y, Ge Z, Li Q, Zhang Y. Circ-ADAM9 targeting PTEN and ATG7 promotes autophagy and apoptosis of diabetic endothelial progenitor cells by sponging mir-20a-5p. Cell Death Dis. 2020;11(7):526.
Jakobi T, Czaja-Hasse LF, Reinhardt R, Dieterich C. Profiling and validation of the circular RNA repertoire in adult murine hearts. Genomics, proteomics and bioinformatics, vol. 14. Beijing Genomics Institute; 2016. p. 216–23.
St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015;31(5):239–51.
Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R, et al. Circular RNAs: Biogenesis, function and role in human diseases. Frontiers in molecular biosciences, vol. 4. Frontiers Media S.A.; 2017. https://pubmed.ncbi.nlm.nih.gov/28634583/
Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–8.
Wang M, Yu F, Li P. Circular RNAs: Characteristics, function and clinical significance in hepatocellular carcinoma. In: Cancers, vol. 10. MDPI AG; 2018. https://pubmed.ncbi.nlm.nih.gov/30072625/
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. CircRNA biogenesis competes with Pre-mRNA splicing. Mol Cell. 2014;56(1):55–66.
Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–34.
Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119(1):440–6.
Liu L, Liu FB, Huang M, Xie K, Xie QS, Liu CH, et al. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int. 2019;18(6):580–6.
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–30.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8.
Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. Cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-Catenin Pathway. PLoS ONE. 2015;10(6):e0131225.
Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268(5209):415–7.
Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66(1):22-37.e9.
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110(3):304–15.
Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5(1):1–9.
Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. Journal of biomedical science, vol. 26. BioMed Central Ltd.; 2019. p. 1–12. Doi: https://doi.org/10.1186/s12929-019-0523-z
Xiao H, Liu M. Circular RNA hsa_circ_0053277 promotes the development of colorectal cancer by upregulating matrix metallopeptidase 14 via miR-2467–3p sequestration. J Cell Physiol. 2020;235(3):2881–90.
Li XN, Wang ZJ, Ye CX, Zhao BC, Huang XX, Yang L. Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed Pharmacother. 2019;112:108611.
Liang Y, Shi J, He Q, Sun G, Gao L, Ye J, et al. Hsa_circ_0026416 promotes proliferation and migration in colorectal cancer via miR-346/NFIB axis. Cancer Cell Int. 2020;20(1):1–15.
Zhang X, Xu Y, Yamaguchi K, Hu J, Zhang L, Wang J, et al. Circular RNA circVAPA knockdown suppresses colorectal cancer cell growth process by regulating miR-125a/CREB5 axis. Cancer Cell Int. 2020;20(1):1–11.
Jin C, Wang A, Liu L, Wang G, Li G. Hsa_circ_0136666 promotes the proliferation and invasion of colorectal cancer through miR-136/SH2B1 axis. J Cell Physiol. 2019;234(5):7247–56.
Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8(12):16020–5.
Geng Y, Zheng X, Hu W, Wang Q, Xu Y, He W, et al. Hsa circ 0009361 acts as the sponge of miR-582 to suppress colorectal cancer progression by regulating APC2 expression. Clin Sci. 2019;133(10):1197–213.
Li X, Wang J, Zhang C, Lin C, Zhang J, Zhang W, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol. 2018;246(2):166–79.
Yang G, Zhang T, Ye J, Yang J, Chen C, Cai S, et al. Circ-ITGA7 sponges miR-3187-3p to upregulate ASXL1, suppressing colorectal cancer proliferation. Cancer Manag Res. 2019;11:6499–509.
Lu H, Yao B, Wen X, Jia B. FBXW7 circular RNA regulates proliferation, migration and invasion of colorectal carcinoma through NEK2, mTOR, and PTEN signaling pathways in vitro and in vivo. BMC Cancer. 2019;19(1):918.
Lu C, Jiang W, Hui B, Rong D, Fu K, Dong C, et al. The circ_0021977/miR-10b-5p/P21 and P53 regulatory axis suppresses proliferation, migration, and invasion in colorectal cancer. J Cell Physiol. 2020;235(3):2273–85.
Wen T, Wu H, Zhang L, Li K, Xiao X, Zhang L, et al. Circular RNA circ_0007142 regulates cell proliferation, apoptosis, migration and invasion via miR-455–5p/SGK1 axis in colorectal cancer. Anticancer Drugs. 2020;32(1):22–33.
He JH, Li YG, Han ZP, Zhou JB, Chen WM, Lv YB, et al. The CircRNA-ACAP2/Hsa-miR-21–5p/Tiam1 regulatory feedback circuit affects the proliferation, migration, and invasion of colon cancer SW480 cells. Cell Physiol Biochem. 2018;49(4):1539–50.
Han K, Wang FW, Cao CH, Ling H, Chen JW, Chen RX, et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol Cancer. 2020;19(1):1–18.
Yang H, Li X, Meng Q, Sun H, Wu S, Hu W, et al. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer. 2020;19(1):1–15.
Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p-TGF-β1 axis. Mol Cancer. 2020;19(1):1–15.
Jian X, He H, Zhu J, Zhang Q, Zheng Z, Liang X, et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol Cancer. 2020;19(1):20. https://doi.org/10.1186/s12943-020-1134-8.
Chen MS, Lin CH, Huang LY, Qiu XM. Circrna smarcc1 sponges mir-140–3p to regulate cell progression in colorectal cancer. Cancer Manag Res. 2020;12:4899–910.
Yan Y, Su M, Qin B. CircHIPK3 promotes colorectal cancer cells proliferation and metastasis via modulating of miR-1207-5p/FMNL2 signal. Biochem Biophys Res Commun. 2020;524(4):839–46.
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7 article. Cell Death Dis. 2018;9(4):1–15.
Chen HY, Li XN, Ye CX, Chen ZL, Wang ZJ. Circular RNA circHUWE1 is upregulated and promotes cell proliferation, migration and invasion in colorectal cancer by sponging mir-486. Onco Targets Ther. 2020;13:423–34.
Wu HB, Huang SS, Lu CG, Tian SD, Chen M. CircAPLP2 regulates the proliferation and metastasis of colorectal cancer by targeting miR-101-3p to activate the Notch signalling pathway. Am J Transl Res. 2020;12(6):2554–69.
Lu C, Fu L, Qian X, Dou L, Cang S. Knockdown of circular RNA circ-FARSA restricts colorectal cancer cell growth through regulation of miR-330–5p/LASP1 axis. Arch Biochem Biophys. 2020;689:108434.
Zhang L, Dong X, Yan B, Yu W, Shan L. CircAGFG1 drives metastasis and stemness in colorectal cancer by modulating YY1/CTNNB1. Cell Death Dis. 2020;11(7):1–15. https://doi.org/10.1038/s41419-020-2707-6.
Ma Z, Han C, Xia W, Wang S, Li X, Fang P, et al. circ5615 functions as a ceRNA to promote colorectal cancer progression by upregulating TNKS. Cell Death Dis. 2020;11(5):1–14. https://doi.org/10.1038/s41419-020-2514-0.
Chen C, Huang Z, Mo X, Song Y, Li X, Li X, et al. The circular RNA 001971/miR-29c-3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. J Exp Clin Cancer Res. 2020;39(1):1–15.
Yang B, Du K, Yang C, Xiang L, Xu Y, Cao C, et al. CircPRMT5 circular RNA promotes proliferation of colorectal cancer through sponging miR-377 to induce E2F3 expression. J Cell Mol Med. 2020;24(6):3431–7.
Wang X, Tao G, Huang D, Liang S, Zheng D. Circular RNA NOX4 promotes the development of colorectal cancer via the microRNA-485–5p/CKS1B axis. Oncol Rep. 2020;44(5):2009–20.
Du J, Xu J, Chen J, Liu W, Wang P, Ye K. CircRAE1 promotes colorectal cancer cell migration and invasion by modulating miR-338–3p/TYRO3 axis. Cancer Cell Int. 2020;20(1):1–14.
Lai M, Liu G, Li R, Bai H, Zhao J, Xiao P, et al. Hsa_circ_0079662 induces the resistance mechanism of the chemotherapy drug oxaliplatin through the TNF-α pathway in human colon cancer. J Cell Mol Med. 2020;24(9):5021–7.
Li Y, Zang H, Zhang X, Huang G. Circ_0136666 facilitates the progression of colorectal cancer via mir-383/creb1 axis. Cancer Manag Res. 2020;12:6795–806.
Li C, Zhou H. Circular RNA hsa_circRNA_102209 promotes the growth and metastasis of colorectal cancer through miR-761-mediated Ras and Rab interactor 1 signaling. Cancer Med. 2020;9(18):6710–25.
Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539–55.
Zhang K, Li S, Gu D, Xu K, Zheng R, Xin J, et al. Genetic variants in circTUBB interacting with smoking can enhance colorectal cancer risk. Arch Toxicol. 2020;94(1):325–33.
Li Y, Pei F, Cao M. CircRNA_101951 promotes migration and invasion of colorectal cancer cells by regulating the KIF3A-mediated EMT pathway. Exp Ther Med. 2020;19(5):3355–61.
Xie Y, Li J, Li P, Li N, Zhang Y, Binang H, et al. RNA-Seq profiling of serum exosomal circular RNAs reveals Circ-PNN as a potential biomarker for human colorectal cancer. Front Oncol. 2020;10:982.
Zhao H, Chen S, Fu Q. Exosomes from CD133+ cells carrying circ-ABCC1 mediate cell stemness and metastasis in colorectal cancer. J Cell Biochem. 2020;121(5–6):3286–97. https://doi.org/10.1002/jcb.29600.
Zeng W, Liu Y, Li WT, Li Y, Zhu JF. CircFNDC3B sequestrates miR-937–5p to derepress TIMP3 and inhibit colorectal cancer progression. Mol Oncol. 2020;14(11):2960–84.
Wang X, Ren Y, Ma S, Wang S. Circular rna 0060745, a novel circrna, promotes colorectal cancer cell proliferation and metastasis through mir-4736 sponging. Onco Targets Ther. 2020;13:1941–51.
Chen ZL, Li XN, Ye CX, Chen HY, Wang ZJ. Elevated levels of circrunx1 in colorectal cancer promote cell growth and metastasis via MiR-145-5p/IGF1 signalling. Onco Targets Ther. 2020;13:4035–48.
Du J, Zhang L, Ma H, Wang Y, Wang P. Retraction: Lidocaine suppresses cell proliferation and aerobic glycolysis by regulating circHOMER1/miR-138–5p/HEY1 axis in colorectal cancer [retraction] (Cancer Manag Res. 2020;12:5009–5022). In: Cancer management and research, vol. 12. Dove Medical Press Ltd; 2020. p. 7341.
Sun J, Liu J, Zhu Q, Xu F, Kang L, Shi X. Hsa_circ_0001806 acts as a ceRNA to facilitate the stemness of colorectal cancer cells by increasing COL1A1. Onco Targets Ther. 2020;13:6315–27.
Zhao JP, Chen LL. Circular RNA MAT2B induces colorectal cancer proliferation via sponging miR-610, resulting in an increased E2F1 expression. Cancer Manag Res. 2020;12:7107–16.
Wang L, Wu H, Chu F, Zhang L, Xiao X. Knockdown of circ_0000512 inhibits cell proliferation and promotes apoptosis in colorectal cancer by regulating miR-296–5p/RUNX1 axis. Onco Targets Ther. 2020;13:7357–68.
Zheng X, Ma YF, Zhang XR, Li Y, Zhao HH, Han SG. Circ-0056618 promoted cell proliferation, migration and angiogenesis through sponging with miR-206 and upregulating CXCR4 and VEGF-A in colorectal cancer. Eur Rev Med Pharmacol Sci. 2020;24(8):4190–202.
Deng Z, Li X, Wang H, Geng Y, Cai Y, Tang Y, et al. Dysregulation of CircRNA_0001946 contributes to the proliferation and metastasis of colorectal cancer cells by targeting MicroRNA-135a-5p. Front Genet. 2020;11:357.
Du H, He Z, Feng F, Chen D, Zhang L, Bai J, et al. Hsa_circ_0038646 promotes cell proliferation and migration in colorectal cancer via miR-331-3p/GRIK3. Oncol Lett. 2020;20(1):266–74.
Wang Y, Wang H, Zhang J, Chu Z, Liu P, Zhang X, et al. Circ_0007031 serves as a sponge of MiR-760 to regulate the growth and chemoradiotherapy resistance of colorectal cancer via regulating dcp1a. Cancer Manag Res. 2020;12:8465–79.
Chen H, Pei L, Xie P, Guo G. Circ-prkdc contributes to 5-fluorouracil resistance of colorectal cancer cells by regulating mir-375/foxm1 axis and wnt/β-catenin pathway. Onco Targets Ther. 2020;13:5939–53.
Dai J, Zhuang Y, Tang M, Qian Q, Chen JP. CircRNA UBAP2 facilitates the progression of colorectal cancer by regulating miR-199a/VEGFA pathway. Eur Rev Med Pharmacol Sci. 2020;24(15):7963–71.
Liu Y, Li H, Ye X, Ji A, Fu X, Wu H, et al. Hsa_circ_0000231 knockdown inhibits the glycolysis and progression of colorectal cancer cells by regulating miR-502–5p/MYO6 axis. World J Surg Oncol. 2020;18(1):1–13.
Chen J, Yang X, Liu R, Wen C, Wang H, Huang L, et al. Circular RNA GLIS2 promotes colorectal cancer cell motility via activation of the NF-κB pathway. Cell Death Dis. 2020;11(9):1–13.
Lin YC, Yu YS, Lin HH, Hsiao KY. Oxaliplatin-induced DHX9 phosphorylation promotes oncogenic circular RNA CCDC66 expression and development of chemoresistance. Cancers (Basel). 2020;12(3):697.
Feng J, Li Z, Li L, Xie H, Lu Q, He X. Hypoxia-induced circCCDC66 promotes the tumorigenesis of colorectal cancer via the miR-3140/autophagy pathway. Int J Mol Med. 2020;46(6):1973–82.
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77(9):2339–50.
Wang X, Chen Y, Liu W, Liu T, Sun D. Hsa_circ_0128846 promotes tumorigenesis of colorectal cancer by sponging hsa-miR-1184 and releasing AJUBA and inactivating Hippo/YAP signalling. J Cell Mol Med. 2020;24(17):9908–24.
DING DY, WANG D, SHU ZB. Erratum: Hsa_circ_0007534 knockdown represses the development of colorectal cancer cells through regulating miR-613/SLC25A22 axis (European Review for Medical and Pharmacological Sciences (2020) 24:6 (3004–3022) DOI: https://doi.org/10.26355/eurrev_202003_20665). European Review for Medical and Pharmacological Sciences, vol. 25. Verduci Editore s.r.l; 2021. p. 570.
Hu B, Xian Z, Zou Q, Zhang D, Su D, Yao J, et al. CircFAT1 suppresses colorectal cancer development through regulating miR-520b/UHRF1 axis or miR-302c-3p/UHRF1 axis. Cancer Biother Radiopharm. 2021;36(1):45–57. https://doi.org/10.1089/cbr.2019.3291.
Xiao YS, Tong HZ, Yuan XH, Xiong CH, Xu XY, Zeng YF. CircFADS2: a potential prognostic biomarker of colorectal cancer. Exp Biol Med. 2020;245(14):1233–41. https://doi.org/10.1177/1535370220929965.
Hao Q, Zhang Z. Hsa_circRNA_000166 facilitated cell growth and limited apoptosis through targeting miR-326/LASP1 axis in colorectal cancer. Gastroenterol Res Pract. 2020;2020:8834359. https://doi.org/10.1155/2020/8834359.
Chen LY, Zhi Z, Wang L, Zhao YY, Deng M, Liu YH, et al. NSD2 circular RNA promotes metastasis of colorectal cancer by targeting miR-199b-5p-mediated DDR1 and JAG1 signalling. J Pathol. 2019;248(1):103–15.
Zhang ZJ, Zhang YH, Qin XJ, Wang YX, Fu J. Circular RNA circDENND4C facilitates proliferation, migration and glycolysis of colorectal cancer cells through miR-760/GLUT1 axis. Eur Rev Med Pharmacol Sci. 2020;24(5):2387–400.
Zhi X, Zhang J, Cheng Z, Bian L, Qin J. circLgr4 drives colorectal tumorigenesis and invasion through Lgr4-targeting peptide. Int J Cancer. 2019. https://doi.org/10.1002/ijc.32549.
Xu XW, Zheng BA, Hu ZM, Qian ZY, Huang CJ, Liu XQ, et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8(53):91674–83.
Li W, Xu Y, Wang X, Cao G, Bu W, Wang X, et al. CircCCT3 modulates vascular endothelial growth factor A and Wnt signaling to enhance colorectal cancer metastasis through sponging miR-613. DNA Cell Biol. 2020;39(1):118–25.
Zhou C, Liu HS, Wang FW, Hu T, Liang ZX, Lan N, et al. circCAMSAP1 promotes tumor growth in colorectal cancer via the miR-328-5p/E2F1 axis. Mol Ther. 2020;28(3):914–28.
He JH, Han ZP, Luo JG, Jiang JW, Zhou JB, Chen WM, et al. Hsa_Circ_0007843 acts as a mIR-518c-5p sponge to regulate the migration and invasion of colon cancer SW480 cells. Front Genet. 2020;11:1. https://doi.org/10.3389/fgene.2020.00009/full.
Zhang J, Wang H, Wu K, Zhan F, Zeng H. Dysregulated circRNA_100876 contributes to proliferation and metastasis of colorectal cancer by targeting microRNA-516b (miR-516b). Cancer Biol Ther. 2020;21(8):733–40.
Xu H, Liu Y, Cheng P, Wang C, Liu Y, Zhou W, et al. CircRNA_0000392 promotes colorectal cancer progression through the miR-193a-5p/PIK3R3/AKT axis. J Exp Clin Cancer Res. 2020;39(1):1–17.
Wu M, Kong C, Cai M, Huang W, Chen Y, Wang B, et al. Hsa_circRNA_002144 promotes growth and metastasis of colorectal cancer through regulating miR-615–5p/LARP1/mTOR pathway. Carcinogenesis. 2020;42(4):601–10.
Chen LY, Wang L, Ren YX, Pang Z, Liu Y, Sun XD, et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138–5p/4EBP-1 mediated cap-independent HIF-1α translation. Mol Cancer. 2020;19(1):1–17.
Liu K, Mou Y, Shi X, Liu T, Chen Z, Zuo X. Circular RNA 100146 promotes colorectal cancer progression by the MicroRNA 149/HMGA2 Axis. Mol Cell Biol. 2020;41(2):e00445-20.
Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N 6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10(1):1–15.
Pei FL, Cao MZ, Li YF. Circ_0000218 plays a carcinogenic role in colorectal cancer progression by regulating miR-139–3p/RAB1A axis. J Infect Dis. 2019;220(4):55–65.
Li Y, Li C, Xu R, Wang Y, Li D, Zhang B. A novel circFMN2 promotes tumor proliferation in CRC by regulating the miR-1182/hTERT signaling pathways. Clin Sci. 2019;133(24):2463–79.
Abu N, Hon KW, Jeyaraman S, Yahaya A, Abdullah NMA, Mustangin M, et al. Identification of differentially expressed circular RNAs in chemoresistant colorectal cancer. Epigenomics. 2019;11(8):875–84.
Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R, et al. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. 2019;30(1):157-173.e7.
Li R, Wu B, Xia J, Ye L, Yang X. Circular RNA hsa_circRNA_102958 promotes tumorigenesis of colorectal cancer via miR-585/CDC25B axis. Cancer Manag Res. 2019;11:6887–93.
Chen Z, Ren R, Wan D, Wang Y, Xue X, Jiang M, et al. Hsa_circ_101555 functions as a competing endogenous RNA of miR-597–5p to promote colorectal cancer progression. Oncogene. 2019;38(32):6017–34.
Lu X, Yu Y, Liao F, Tan S. Homo sapiens circular RNA 0079993 (hsa-circ-0079993) of the POLR2J4 gene acts as an oncogene in colorectal cancer through the microRNA-203a-3p.1 and CREB1 axis. Med Sci Monit. 2019;25:6872–83.
Zhang Q, Zhang C, Ma JX, Ren H, Sun Y, Xu JZ. Circular rna pip5k1a promotes colon cancer development through inhibiting mir-1273a. World J Gastroenterol. 2019;25(35):5300–9.
Zhang J, Liu H, Zhao P, Zhou H, Mao T. Has_circ_0055625 from circRNA profile increases colon cancer cell growth by sponging miR-106b-5p. J Cell Biochem. 2019;120(3):3027–37.
Yong W, Zhuoqi X, Baocheng W, Dongsheng Z, Chuan Z, Yueming S. Hsa_circ_0071589 promotes carcinogenesis via the miR-600/EZH2 axis in colorectal cancer. Biomed Pharmacother. 2018;102:1188–94.
Chen L, Zhang S, Wu J, Cui J, Zhong L, Zeng L, et al. CircRNA-100290 plays a role in oral cancer by functioning as a sponge of the MIR-29 family. In: Oncogene, vol. 36. Nature Publishing Group; 2017. p. 4551–61.
Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, et al. Circular RNA ciRS-7—a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23(14):3918–28.
Xiong W, Ai YQ, Li YF, Ye Q, Chen ZT, Qin JY, Liu QY, Wang H, Ju YH, Li WH, Li YF. Microarray analysis of circular RNA expression profile associated with 5-fluorouracil-based chemoradiation resistance in colorectal cancer cells. Biomed Res Int. 2017;2017:8421614. https://doi.org/10.1155/2017/8421614.
Han J, Zhao G, Ma X, Dong Q, Zhang H, Wang Y, et al. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun. 2018;503(4):2429–35.
Wang L, Peng X, Lu X, Wei Q, Chen M, Liu L. Inhibition of hsa_circ_0001313 (circCCDC66) induction enhances the radio-sensitivity of colon cancer cells via tumor suppressor miR-338–3p: Effects of cicr_0001313 on colon cancer radio-sensitivity. Pathol Res Pract. 2019;215(4):689–96.
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–91.
Huang X, Shen X, Peng L, Mai W, Wang Y, Zheng H. CircCSNK1G1 contributes to the development of colorectal cancer by increasing the expression of MYO6 via competitively targeting miR-455–3p. Cancer Manag Res. 2020;12:9563–75.
Li Z, Yao H, Wang S, Li G, Gu X. CircTADA2A suppresses the progression of colorectal cancer via miR-374a-3p/KLF14 axis. J Exp Clin Cancer Res. 2020;39(1):1–14.
Wang DK, Chong RF, Song BL, Fan KF, Liu YF. Circular RNA circ-SMAD7 is downregulated in colorectal cancer and suppresses tumor metastasis by regulating epithelial mesenchymal transition. Eur Rev Med Pharmacol Sci. 2020;24(4):1736–42.
Xu B, Yang N, Liu Y, Kong P, Han M, Li B. Circ_cse1l inhibits colorectal cancer proliferation by binding to eIF4A3. Med Sci Monit. 2020;26:e923876-1.
Huang G, Ma J, Zhang L. Integrin subunit alpha 5 (ITGA5) gene circular rna sponges microrna-107 in colorectal carcinoma cells and tissues and regulates the expression of the forkhead box J3 (FOXJ3) gene. Med Sci Monit. 2020;26:e920623-1.
Ren TJ, Liu C, Hou JF, Shan FX. CircDDX17 reduces 5-fluorouracil resistance and hinders tumorigenesis in colorectal cancer by regulating miR-31–5p/KANK1 axis. Eur Rev Med Pharmacol Sci. 2020;24(4):1743–54.
Yang Z, Zhang J, Lu D, Sun Y, Zhao X, Wang X, et al. Hsa_circ_0137008 suppresses the malignant phenotype in colorectal cancer by acting as a microRNA-338–5p sponge. Cancer Cell Int. 2020;20(1):1–12.
Zhang Y, Zhang Z, Yi Y, Wang Y, Fu J. CircNOL10 acts as a sponge of miR-135a/b-5p in suppressing colorectal cancer progression via regulating KLF9. Onco Targets Ther. 2020;13:5165–76.
Li H, Jin X, Liu B, Zhang P, Chen W, Li Q. CircRNA CBL.11 suppresses cell proliferation by sponging miR-6778–5p in colorectal cancer. BMC Cancer. 2019;19(1):1–14.
Cui W, Dai J, Ma J, Gu H. Circcdyl/microRNA-150–5p participates in modulating growth and migration of colon cancer cells. Gen Physiol Biophys. 2019;38(6):485–95.
Jin Y, Yu LL, Zhang B, Liu CF, Chen Y. Circular RNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge. Braz J Med Biol Res. 2018;51(12).
Raza U, Zhang JD, Şahin Ö. MicroRNAs: Master regulators of drug resistance, stemness, and metastasis. In: Journal of molecular medicine, vol. 92. Springer Verlag; 2014. p. 321–36.
Wr J, Ne S. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–61.
Jin Y, Yu LL, Zhang B, Liu CF, Chen Y. Circular RNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge. Braz J Med Biol Res. 2018;51(12):e7811. https://doi.org/10.1590/1414-431X20187811.
Yang N, Xu B, Kong P, Han M, Li BH. Hsa_circ_0002320: a novel clinical biomarker for colorectal cancer prognosis. Medicine (Baltimore). 2020;99(28):e21224.
Tang X, Sun G, He Q, Wang C, Shi J, Gao L, et al. Circular noncoding RNA circMBOAT2 is a novel tumor marker and regulates proliferation/migration by sponging miR-519d-3p in colorectal cancer. Cell Death Dis. 2020;11(8):1–15.
Sadeghi H, Heiat M. A novel circular RNA hsa_circ_0060927 may serve as a potential diagnostic biomarker for human colorectal cancer. Mol Biol Rep. 2020;47(9):6649–55.
Jin YD, Ren YR, Gao YX, Zhang L, Ding Z. Hsa_circ_0005075 predicts a poor prognosis and acts as an oncogene in colorectal cancer via activating Wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci. 2019;23(8):3311–9.
Xing L, Xia M, Jiao X, Fan L. Hsa_circ_0004831 serves as a blood-based prognostic biomarker for colorectal cancer and its potentially circRNA-miRNA-mRNA regulatory network construction. Cancer Cell Int. 2020;20(1):1–9.
Min L, Wang H, Zeng Y. CircRNA-104916 regulates migration, apoptosis and epithelial-mesenchymal transition in colon cancer cells. Front Biosci Landmark. 2019;24:819–32. https://doi.org/10.2741/4753.
Tian J, Xi X, Wang J, Yu J, Huang Q, Ma R, et al. CircRNA hsa_circ_0004585 as a potential biomarker for colorectal cancer. Cancer Manag Res. 2019;11:5413–23.
Pan B, Qin J, Liu X, He B, Wang X, Pan Y, et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096.
Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18(1):1–13.
Gi Z, Li LF, Wang CY, Wang Y, Ma WL. CircMTO1 inhibits cell proliferation and invasion by regulating Wnt/β-catenin signaling pathway in colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22(23):8203–9.
Ji W, Qiu C, Wang M, Mao N, Wu S, Dai Y. Hsa_circ_0001649: A circular RNA and potential novel biomarker for colorectal cancer. Biochem Biophys Res Commun. 2018;497(1):122–6.
Wang F, Wang J, Cao X, Xu L, Chen L. Hsa_circ_0014717 is downregulated in colorectal cancer and inhibits tumor growth by promoting p16 expression. Biomed Pharmacother. 2018;98:775–82.
Li J, Ni S, Zhou C, Ye M. The expression profile and clinical application potential of hsa_circ_0000711 in colorectal cancer. Cancer Manag Res. 2018;10:2777–84.
Wang J, Li X, Lu L, He L, Hu H, Xu Z. Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J Clin Lab Anal. 2018;32(5):e22379.
Zhou F, Lin H, Chen Z, Huang Z, Hu J. The expression profile and clinical significance of circRNA0003906 in colorectal cancer. Onco Targets Ther. 2017;10:5187–93.
Sang M, Meng L, Liu S, Ding P, Chang S, Ju Y, et al. Circular RNA ciRS-7 maintains metastatic phenotypes as a ceRNA of miR-1299 to target MMPs. Mol Cancer Res. 2018;16(11):1665–75.
Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. In: Nature materials, vol. 12. Nature Publishing Group; 2013. p. 967–77.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
MA and MA contributed equally in conception, design and data collection and drafting of the manuscript. C.Y and M.H contributed in editing and drafting the manuscript and E. N and M. B were the supervising and revising the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest, All authors approved the final version for submission.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ameli-Mojarad, M., Ameli-Mojarad, M., Hadizadeh, M. et al. The effective function of circular RNA in colorectal cancer. Cancer Cell Int 21, 496 (2021). https://doi.org/10.1186/s12935-021-02196-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-021-02196-0