Abstract
Background
Long noncoding RNAs (lncRNAs) have been elucidated to participate in the development and progression of various cancers. In this study, we aimed to explore the underlying functions and mechanisms of LINC00958 in colorectal cancer.
Methods
LINC00958 expression in colorectal cancer tissues was examined by qRT-PCR. The correlations between LINC00958 expression and clinical characteristics and prognosis were evaluated. The biological functions of LINC00958 were detected by CCK-8, MTT, colony formation and flow cytometric analyses. RNA pulldown, RIP and luciferase reporter assays were used to confirm the regulatory effects of LINC00958 on miR-422a. Rescue experiments were performed to detect the effects of miR-422a on the roles of LINC00958.
Results
LINC00958 was upregulated in colorectal cancer tissues and cell lines. High LINC00958 levels were positively associated with T stage and predicted poor prognosis. Cell experiments showed that LINC00958 promoted cell proliferation and suppressed apoptosis and sensitivity to radiotherapy in vitro and promoted tumor growth in vivo. Bioinformatics analysis predicted the binding site of miR-422a on LINC00958. Mechanistically, RNA pulldown, RIP and luciferase reporter assays demonstrated that LINC00958 specifically targeted miR-422a. In addition, we found that miR-422a suppressed MAPK1 expression by directly binding to the 3’-UTR of MAPK1, thereby inhibiting cell proliferation and enhancing cell apoptosis and radiosensitivity. Furthermore, miR-422a rescued the roles of LINC00958 in promoting MAPK1 expression and cell proliferation and decreasing cell apoptosis and radiosensitivity.
Conclusions
LINC00958 promoted MAPK1 expression and cell proliferation and suppressed cell apoptosis and radiosensitivity by targeting miR-422a, which suggests that it is a potential biomarker for the prognosis and treatment of colorectal cancer.
Similar content being viewed by others
Introduction
Colorectal cancer is one of the most serious malignancies and the second main cause of cancer-related death worldwide [1, 2]. With advancements in early diagnosis and combined treatments, the morbidity and mortality of colorectal cancer patients aged 65 years and older have declined by 3.3% and 3.0% annually, respectively [3]. However, the incidence and death rates in individuals younger than 50 years have increased by 1% and 1.3% annually [3], respectively. To date, the potential molecular mechanisms underlying the development and progression of colorectal cancer remain ambiguous. Thus, it is urgent to explore the molecular mechanisms and identify more effective molecular targets for the early diagnosis and treatment of colorectal cancer.
Human genomes produce a great quantity of noncoding RNAs with limited protein coding potential, many of which participate in diverse biological processes. Recently, long noncoding RNAs (lncRNAs), which are larger than 200 nucleotides in length, have attracted great attention [4, 5]. Dysregulation of lncRNAs has occurred in various types of cancer. Increasing evidence has shown that lncRNAs play vital roles in the pathogenesis and progression of cancers, participating in processes such as cell proliferation, apoptosis, angiogenesis, lymphangiogenesis, cell signaling transduction and distant metastasis [6,7,8,9]. Pan et al. [10] revealed that the lncRNA FOXC2-AS1 was upregulated in colorectal cancer tissues, and si-FOXC2-AS1 suppressed cell proliferation, invasion and metastasis in vitro and in vivo. In addition, Tian et al. identified a novel lncRNA, GCMA, which was highly expressed in gastric cancer tissues and predicted poor prognosis. Additionally, they demonstrated that GCMA promoted cell proliferation, epithelial-mesenchymal transition (EMT) and cell metastasis [11]. Adjuvant radiotherapy or palliative radiotherapy contributes to downstaging or delaying tumor progression in many types of cancer [12]. Successful radiotherapy is based on a great understanding of the radiotherapy mechanisms. Recently, Wang et al. revealed that the lncRNA CCAT2 suppressed cell apoptosis and radiosensitivity in human esophageal carcinoma [13]. However, the accurate mechanisms of lncRNAs in the progression of cancers have not been elucidated. Currently, lncRNAs have been found to exert their biological functions in four different ways, namely, as signals, decoys, guides and scaffolds [14]. Wu et al. reported that the lncRNA PVT1 acted as a miRNA sponge to relieve the inhibition of VEGFA by miR-16-5p and activated the VEGFA/VEGFR/AKT pathway, thereby promoting the tumorigenesis of colorectal cancer [15]. Hua et al. [16] showed that the hypoxia-induced lncRNA AC020978 promoted cell proliferation and glycolytic metabolism in non-small cell lung cancer by directly interacting with PKM2 and enhancing the stability of the PKM2 protein. As the most important mechanism, the miRNA sponge mechanism has been a research hotspot of lncRNAs in recent years.
In this study, we found that the lncRNA LINC00958 was upregulated in colorectal cancer tissues and cell lines and significantly associated with clinicopathological features and prognosis. As the most important member of the mitogen-activated protein kinase family, MAPK1 is widely accepted to play oncogenic roles in various cancer types, such as cell proliferation, apoptosis and radiosensitivity [17,18,19,20]. We demonstrated that LINC00958 served as a miR-422a sponge and enhanced MAPK1 expression, thereby promoting cell proliferation and suppressing apoptosis and radiosensitivity. Our data suggest that the LINC00958/miR-422a/MAPK1 axis plays key roles in the cell proliferation and radiosensitivity of colorectal cancer and may be a promising candidate in the diagnosis and treatment of colorectal cancer.
Materials and methods
Tissues samples and cell culture
From 2013 to 2018, 63 pairs of fresh frozen samples of colorectal cancer tissues and adjacent normal tissues were collected from colorectal cancer patients in Henan Provence People’s Hospital. All the samples were snap-frozen into an RNA keeper tissue stabilizer (Vazyme Biotech Co., Jiangsu, China) and kept in a -80 °C freezer after storage at 4 °C overnight. All patients underwent radical resection without radiotherapy or chemotherapy before surgery. Written informed consent was acquired from all the subjects. The study was approved by the Institutional Review Board of Henan Provence People’s Hospital.
Human colorectal cancer cells and human normal colorectal mucosa FHC cells were purchased from the Type Culture Collection of the Chinese Academy of Science (Shanghai, China). All the cells were cultured in DMEM (HyClone, USA) with 10% fetal bovine serum (Gibco, Australia) and 1% penicillin–streptomycin in a humidified atmosphere containing 5% CO2 at 37 °C.
Transfection and oligonucleotides and plasmids
To regulate the expression of LINC00958 and miR-422a, siRNA targeting LINC00958 (si-LINC00958: 5’-GTGACTAGCTTAAACTAAATT-3’) and an overexpression plasmid were synthesized; the miR-422a mimics and inhibitor were purchased from RiboBio (Guangzhou, China). When colorectal cancer cells grew to 80% confluence, they were digested with 0.25% trypsin, resuspended and seeded in 6-well plates. According to the manufacturer’s instructions, the plasmids or oligonucleotides were transfected into cells with LipofectamineTM 2000 (Invitrogen, USA) after culturing for 24 h.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cells and tissues with TRIzol (TaKaRa, Shiga, Japan) according to the manufacturer’s instructions. RNA was reverse transcribed into cDNA using the PrimeScript™ RT Master Mix Reagent Kit (TaKaRa, Shiga, Japan) for lncRNA and mRNA, and cDNA was synthesized by the PrimeScript™ RT Reagent Kit (TaKaRa, Shiga, Japan) for miRNA. Then, qRT-PCR was conducted to quantify RNA levels with SYBR Premix Ex Taq™ (TaKaRa, Shiga, Japan). GAPDH or U6 was used as an internal control. The primers were as follows: LINC00958, F: 5’-CCATTGAAGATACCACGCTGC-3’, R: 5’-GGTT GTTGCCCAGGGTAGTG-3’; MAPK1, F: 5’-CTGGACGTGCTCAGACATCG-3’, R: 5’-GGTCAGCAGGGCATC ATGTAG-3’; and GAPDH, F: 5’-CACCATTGGCAATGAGCGGTTC-3’, R: 5’-AGGTC TTTGCGGATGTCCACGT-3’. The relative expression levels of RNAs were calculated by the ΔΔCt method.
CCK-8 and MTT assays
A total of 2000 cells were seeded into 96-well plates and cultured at 37 °C. After culturing for 0 h, 24 h, 48 h, 72 h and 96 h, the optical density of each well was measured with a CCK-8 kit or an MTT kit (Beyotime Biotechnology, Jiangsu, China) at 450 nm or 570 nm. All experiments were executed in triplicate.
Flow cytometric analysis
Cell apoptosis assays were conducted with the Annexin V Apoptosis Detection Kit (FITC) (eBioscience, USA). Cells were treated with trypsin without EDTA. Then, the cells were washed with precooled PBS and diluted with 100 μl of binding buffer. Then, 5 μl of fluorochrome-conjugated Annexin V was added and incubated at room temperature for 15 min. After resuspension in 200 μl of binding buffer, 5 μl of PI was added. Finally, the cell apoptosis percentages were detected and analyzed by flow cytometry. All experiments were executed in triplicate.
Radiation exposure and colony formation assay
A single dose of ionizing radiation was delivered by a Siemens 6 MV X-ray linear accelerator with a distance of 100 cm from the source skin at a dose rate of 2 Gy/min at room temperature. Two hundred transfected SW480 or HCT8 cells were seeded in six-well plates. Five dishes of cells were irradiated for 8 h with different gradient irradiation doses of 0, 2, 4, 6, and 8 Gy. After incubation for 14 days at 37 °C, the cells were fixed with 75% ethanol and stained with 0.1% crystalline purple. The number of colonies containing 50 cells or more was counted under an inverted microscope (40 × magnification, Leica, Germany). The relative survival fraction was calculated as the ratio of plating efficiency (treated) to PE (control): PE = number of colonies/number of seeded cells; SF = PE (irradiated cells)/PE (control cells). All experiments were executed in triplicate.
Western blotting
Total protein was extracted from colorectal cancer cells using radioimmunoprecipitation assay (RIPA) buffer (Beyotime Biotechnology, Jiangsu, China) according to the manufacturer’s instructions. Protein concentrations were tested with a BCA protein assay kit (Beyotime Biotechnology, Jiangsu, China). Total protein (30 μg) was separated by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, MA, USA). The membranes were blocked with 5% nonfat milk for 1.5 h at room temperature and then incubated with primary antibodies at 4 °C overnight. After incubation with secondary antibodies at room temperature for 1.5 h, protein bands were detected by ECL chemiluminescent reagent (Millipore, MA, USA). The primary antibodies were as follows: Erk1/2 (1:1000; Cell Signaling Technology), pErk (1:1000; Cell Signaling Technology), BCL2 (1:1000; Cell Signaling Technology), Bax (1:2000; Abcam) and GAPDH (1:5000; Cell Signaling Technology).
RNA fluorescence in situ hybridization (FISH) and nuclear-cytoplasmic fractionation
RNA-FISH assays were carried out to confirm the subcellular location of LINC00958 in colorectal cancer cells. The Cy3-labeled LINC00958 probe was synthesized by GenePharma (Shanghai, China). The sequence of the Cy3-labeled LINC00958 probe was as follows: 5’-TCCTCCCATGTTTTTGTCTTCCCTACCACC-3’. Hybridization was performed according to the manufacturer’s instructions. The images were acquired using a fluorescence microscope. Nuclear and cytoplasmic RNA was isolated by a PARIS™ kit (Invitrogen, USA) according to the manufacturer’s instructions and then detected by qRT-PCR.
RNA pull down assay
The biotinylated LINC00958 probe was produced by RiboBio (Guangzhou, China). In brief, 1 × 107 SW480 cells were lysed with ultrasonication. Probe-coated beads were first incubated with the oligo probe or LINC00958 probe using C-1 magnetic beads (Life Technologies) for 2 h at 25 °C. Next, the cell lysates were incubated with C-1 magnetic beads combined with the oligo probe or LINC00958 probe (Biotin-LINC00958 of wild type: 5’-TCCTCCCATGTTTTTGTCTTCCCTACCACC-3’; Bio LINC00958 of mutation: 5’-AGGAGGGTACAAAAACAGAAGGGATGGTGG-3’) at 4 °C overnight. After washing with wash buffer three times, the RNA complexes were extracted by the RNeasy Mini Kit (QIAGEN, Germany) according to the manufacturer’s instructions. Finally, qRT-PCR was conducted to quantify the level of miR-422a.
RNA immunoprecipitation (RIP) assay
The RIP assay was conducted in SW480 cells with the Magna RIPTM RNA-binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA). SW480 cells were transfected with miR-422a mimics or negative control. After transfection for 48 h, the cells were lysed with complete RNA lysis buffer. Magnetic beads were first conjugated with human anti-AGO2 antibody or negative control mouse IgG. Then, cell lysates were rotated in RIP immunoprecipitation buffer with the above magnetic beads. The next day, immunoprecipitated RNA was incubated with Proteinase K for 30 min and extracted by TRIzol. Finally, qRT-PCR was performed to identify the level of LINC00958.
Luciferase reporter assay
Luciferase reporter plasmids (pGL3-LINC00958 sequence, pGL3-mutant LINC00958 sequence, pGL3-MAPK1 3’-UTR sequence and pGL3-mutant MAPK1 3’-UTR sequence) were produced by HarO Life Co. (Shanghai, China). The luciferase reporter plasmids were transfected into cells with the miR-422a mimic or inhibitor. After 36 h, the activities of firefly luciferase and Renilla luciferase were detected. The relative luciferase activity was calculated as firefly luciferase activity/Renilla luciferase activity *100%.
Animal experiments
To establish xenograft tumor models, shRNAs against LINC00958 (sh-LINC00958) and a negative control (sh-NC) were generated and cloned into lentiviruses. Then, lentiviruses were transfected into SW480 cells with 5 mg/ml polybrene for 48 h. Finally, stable SW480 cells were selected with puromycin (5 μg/ml) for 2 weeks. The knockdown efficiency was confirmed by qRT-PCR. For the in vivo tumorigenesis assay, 10 4-week-old male BALB/c athymic nude mice were randomly divided into two groups (n = 5). A total of 1.0 × 107 stable SW480 cells in 150 μl of PBS were subcutaneously injected into the left inguinal region of the mice. After 10 days, tumor volumes were measured every three days until 4 weeks. Tumor volume was calculated by the following formula: tumor volume = (length × width2)/2. Finally, the mice were sacrificed, and the volume and weight of tumors were detected. The animal experiments were approved by the Institutional Animal Care and Use Committee of Zhengzhou University and performed according to the guidelines for the care and use of laboratory animals.
Statistical analysis
SPSS 22.0 software was used for statistical analyses. Count data were analyzed by the χ2 test or Fisher’s exact test. Paired and unpaired measurement data were compared by Student’s t-test or the Mann–Whitney U test. The survival curves were calculated by the Kaplan–Meier method and analyzed by the log-rank test. A probability of 0.05 or less was considered statistically significant for all tests.
Results
LINC00958 is overexpressed in colorectal cancer tissues
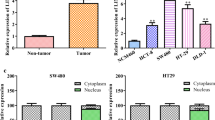
Previous studies showed that the level of LINC00958 was upregulated in cancers, such as hepatocellular carcinoma and oral squamous cell carcinoma, and predicted poor prognosis [21, 22]. In this study, the level of LINC00958 was detected with qRT-PCR technology in 63 paired colorectal cancer tissues and matched adjacent normal tissues. We found that LINC00958 expression was upregulated in colorectal cancer tissues compared with matched normal tissues (82.54%, 52/63) (Fig. 1a, b). Next, the correlations between the level of LINC00958 and clinicopathological features were analyzed in these 63 paired colorectal cancer tissues. The results revealed that the level of LINC00958 was positively correlated with tumor differentiation, T stage and TNM stage (Table 1, P < 0.05). The level of LINC00958 was remarkably higher for stage T3 and T4 disease than for stage T1 and T2 disease (Fig. 1c). However, there were no significant differences in colorectal cancer tissues between different differentiation and TNM stages. Kaplan–Meier survival analysis indicated that colorectal cancer patients with higher LINC00958 expression had worse overall survival (Fig. 1d) and disease-free survival (Fig. 1e) than those with lower LINC00958 expression. Overall, these results show that LINC00958 is highly expressed in colorectal cancer tissues and may act as a promising diagnostic, prognostic and therapeutic marker for colorectal cancer.
Features of LINC00958 in colorectal cancer tissues. a LINC00958 expression was upregulated in most (82.54%, 52/63) of colorectal cancer tissues. b LINC00958 expression was significantly higher in 63 colorectal cancer tissues than adjacent normal colorectal tissues. c The level of LINC00958 was significantly higher in colorectal cancer tissues at T3 and T4 stages than these at T1 and T2 stages. d and e Kaplan–Meier Plotter analysis revealed that high LINC00958 level predicted a poorer overall survival (d) and disease-free survival (e) for colorectal cancer patients. *p < 0.05, **p < 0.01, NSp > 0.05
LINC00958 promotes cell proliferation and suppresses cell apoptosis and the radiosensitization of colorectal cancer in vitro
The expression of LINC00958 in colorectal cancer cell lines was detected using qRT-PCR. The results showed that the level of LINC00958 was higher in colorectal cancer cell lines than in FHC human normal colorectal mucosa cells (Fig. 2a). On the basis of the expression of LINC00958, SW480 and HCT8 cells were selected to further investigate the function of LINC00958. Then, siRNA targeting LINC00958 and an LINC00958 overexpression plasmid were designed and synthesized. The qRT-PCR results showed that si-LINC00958 markedly decreased the expression of LINC00958 in SW480 cells and that the overexpression plasmid significantly increased its expression in HCT8 cells (Fig. 2b). To detect the roles of LINC00958 in colorectal cancer cells, flow cytometry, CCK-8, MTT and colony formation assays were performed under different radiation doses. The results of the flow cytometry assay showed that knockdown of LINC00958 significantly increased the cell apoptosis percentage (Fig. 2c), while overexpression of LINC00958 remarkably decreased the cell apoptosis percentage (Fig. 2g). The CCK-8 and MTT results indicated that si-LINC00958 decreased cell proliferation (Fig. 2d, e), while overexpression of LINC00958 increased cell proliferation (Fig. 2h, i). In addition, we found that downregulation of LINC00958 decreased the survival fraction under different irradiation doses (Fig. 2f), and upregulation of LINC00958 increased the survival fraction under different irradiation doses (Fig. 2j), indicating that LINC00958 decreased the radiosensitization of colorectal cancer cells. Taken together, these results demonstrated that LINC00958 promoted cell proliferation and suppressed apoptosis and the radiosensitivity of colorectal cancer cells.
LINC00958 promotes cell proliferation, suppresses cell apoptosis and radiosensitization of colorectal cancer. a The expression of LINC00958 in colorectal cancer cell lines. b The efficiencies of siRNA targeting LINC00958 and an overexpression plasmid of LINC00958 were detected. c The flow cytometry assay showed that knockdown of LINC00958 significantly increased the percentage of apoptosis. d, e The CCK-8 (d) and MTT (e) results indicated that si-LINC00958 decreased the ability of cell proliferation. f si-LINC00958 decreased the survival fraction under different irradiation dose. g The flow cytometry assay showed that overexpression of LINC00958 remarkably decreased the percentage of apoptosis. h and i The CCK-8 (h) and MTT (i) results indicated that overexpression of LINC00958 increased the ability of cell proliferation. j Overexpression of LINC00958 increased the survival fraction under different irradiation dose. Data are reported as means ± standard deviation of three independent experiments. *p < 0.05, **p < 0.01
LINC00958 promotes colorectal cancer cell proliferation in vivo
To explore the roles of LINC00958 in colorectal cancer growth in vivo, stable SW480 cells (transfected with sh-LINC00958 or sh-NC) and a nude mouse xenograft model were constructed. The knockdown efficiency was detected by qRT-PCR. The results showed that sh-LINC00958 significantly decreased the expression of LINC00958 in SW480 cells (Fig. 3a). Representative images of SW480 cell tumor formation are shown in Fig. 3b. The weight of the SW480/sh-LINC00958 tumors was significantly lower than that of the SW480/sh-NC tumors (Fig. 3c). In addition, the volume of the SW480/sh-LINC00958 tumors was remarkably lower than that of the SW480/sh-NC tumors (Fig. 3d). Collectively, our results demonstrate that LINC00958 promotes colorectal cancer cell proliferation in vivo.
LINC00958 promotes colorectal cancer cell proliferation in vivo. a The knockdown efficiency of stable SW480/sh-LINC00958 cells was detected by qRT-PCR. b Represent pictures of tumor formation of SW480 cells. c The final tumor weight of SW480 cells was shown. d Tumor volumes of SW480 cells were measured every three days. **p < 0.01
LINC00958 acts as a miRNA sponge of miR-422a in colorectal cancer cells
Since lncRNAs exert biological functions mainly by acting as miRNA sponges, we explored whether LINC00958 promotes colorectal cancer progression by sponging miRNAs. To explore the potential target miRNAs of LINC00958, the TargetScan (http://www.targetscan.org) and StarBase (http://starbase.sysu.edu.cn) databases were employed to predict the underlying miRNAs and binding sites of LINC00958. Both databases showed that LINC00958 contained the binding site of miR-422a (Fig. 4a). The qRT-PCR results showed that si-LINC00958 significantly increased the expression of miR-422a in SW480 cells, and overexpression of LINC00958 decreased miR-422a expression (Fig. 4b), implying a potential correlation between LINC00958 and miR-422a. To analyze the correlation between LINC00958 and miR-422a, the level of miR-422a in colorectal cancer tissues was detected. The results showed that miR-422a expression was downregulated in most colorectal cancer tissues (74.60%, 47/63) (Fig. 4c). Further analysis indicated that the level of LINC00958 was negatively correlated with the level of miR-422a in colorectal cancer tissues (R = -0.5122, P < 0.01) (Fig. 4d). To identify the subcellular location of LINC00958 in colorectal cancer cells, RNA-FISH assays were conducted with the Cy3-labeled LINC00958 probe. The results showed that LINC00958 was mostly located in the cytoplasm of SW480 cells (Fig. 4e). In addition, nuclear-cytoplasmic fractionation assays were performed. The results showed that LINC00958 was mostly located in the cytoplasm (Fig. 4f). Considering that the location of lncRNAs impacts their functions, we speculated that LINC00958 might affect gene expression as a competitive endogenous RNA (ceRNA). Next, biotinylated RNA pulldown and RIP assays were performed to identify whether LINC00958 directly interacted with miR-422a. The RNA pulldown assay showed that miR-422a was captured by wild-type LINC00958 but not mutant LINC00958 (Fig. 4g). The RIP assay was conducted to immunoprecipitate LINC00958 with an anti-AGO2 antibody or control IgG. The results showed that LINC00958 was enriched by miR-422a mimics compared with the negative control (Fig. 4h). These data demonstrated that LINC00958 directly interacted with miR-422a. Furthermore, to investigate the regulatory effects of LINC00958 on miR-422a, we constructed two luciferase reporter plasmids with wild-type LINC00958 (WT) and mutated LINC00958 (MUT) in which the binding site of LINC00958 on miR-422a was mutated (Fig. 4a). Luciferase reporter assays showed that miR-422a mimics significantly decreased the luciferase activity of wild-type LINC00958 but not mutant LINC00958 (Fig. 4i), while the miR-422a inhibitor evidently increased the luciferase activity of wild-type LINC00958 (Fig. 4j). In general, these data demonstrated that LINC00958 acted as a miR-422a sponge by directly binding to MREs.
LINC00958 acts as a miRNA sponge of miR-422a. a Bioinformatics databases predicted that LINC00958 contained the binding site of miR-422a. b Expression of miR-422a after knockdown or overexpression of LINC00958 in colorectal cancer cells. c The expression of miR-422a in 63 paired colorectal cancer tissues. d Pearson’s correlation showed that miR-422a level negatively correlated with LINC00958 level in 63 paired colorectal cancer tissues (R = − 0.5122, p < 0.001). e RNA-FISH assays showed that most of LINC00958 was located in the cytoplasm of SW480 cells. f qRT-PCR results of U6, GAPDH and LINC00958 expressions in cell nucleus and cytoplasm. g The biotinylated RNA pull-down showed that miR-422a was pulled down by LINC00958 probe, but not by mutant LINC00958 probe, in SW480 cells. h RIP assay was performed with AGO2 antibody in SW480 cells transfected with miR-422a mimics or NC, and the enrichment of LINC00958 was detected. i miR-422a mimics significantly decreased the luciferase activity of wild type of LINC00958, but not the activity of mutation of LINC00958. j miR-422a inhibitor evidently increased the luciferase activity of wild type of LINC00958, but not it of mutation of LINC00958. Three independent experiments were performed for each group. All data are reported as the mean ± SD. **p < 0.01
MiR-422a suppresses cell proliferation and increases cell apoptosis and the radiosensitization of colorectal cancer through MAPK1
To explore the potential target genes of miR-422a, the miRDB (http://mirdb.org), TargetScan and miRanda (http://www.miranda.org) databases were employed to predict underlying binding sites. All three databases predicted that the MAPK1 3’-untranslated region (3’-UTR) contained the binding site of miR-422a (Fig. 5a). Next, we explored whether MAPK1 is a target gene of miR-422a. The qRT-PCR results showed that miR-422a mimics markedly decreased the expression of MAPK1, while the miR-422a inhibitor significantly increased its expression (Fig. 5b). The Western blot analysis results indicated that overexpression of miR-422a decreased the expression of both ERK1/2 and p-ERK1/2 (MAPK1 protein), decreased the expression of Bcl-2 and increased the expression of Bax, the downstream protein of the ERK pathway, while knockdown of miR-422a increased the expression of ERK1/2, p-ERK1/2 and Bcl-2 but decreased the expression of Bax (Fig. 5c). To explore the regulatory mechanisms of miR-422a on MAPK1, luciferase reporter plasmids with wild-type MAPK1 mRNA 3’-UTR (WT) or mutant MAPK1 mRNA 3’-UTR (MUT) with mutant binding sites of MAPK1 on miR-422a were constructed (Fig. 5a). The luciferase reporter assay results implied that overexpression of miR-422a decreased the luciferase activity of WT but did not decrease that of MUT, while knockdown of miR-422a remarkably increased the luciferase activity of WT but not that of MUT (Fig. 5d). In summary, these data proved that miR-422a negatively regulated MAPK1 expression by directly binding to the MAPK1 3’-UTR.
miR-422a suppresses cell proliferation, increases cell apoptosis and radiosensitization of colorectal cancer through MAPK1. a Bioinformatics databases predicted that MAPK1 was a target of miR-422a. b, c mRNA and protein expression of MAPK1 after knockdown or overexpression of miR-422a in colorectal cancer cells. d miR-422a mimics significantly decreased the luciferase activity of wild type of MAPK1, but not the activity of mutation of MAPK1. In addition, miR-422a inhibitor evidently increased the luciferase activity of wild type of MAPK1, but not it of mutation of MAPK1. e, i The flow cytometry assay showed that miR-422a mimics significantly increased the apoptosis percentage (e), while miR-422a inhibitor remarkably decreased the apoptosis percentage (i). f, g) and j, k The CCK-8 and MTT results indicated that miR-422a mimics decreased the ability of cell proliferation, and miR-422a inhibitor increased it. h, l miR-422a mimics enhanced the radiosensitization of colorectal cancer cell (h), while miR-422a inhibitor decreased the radiosensitization (l). Data are reported as means ± standard deviation of three independent experiments. **p < 0.01
To test the effects of miR-422a on colorectal cancer cells, flow cytometry, CCK-8, MTT and colony formation assays were performed under different radiation doses. The flow cytometry assay showed that miR-422a mimics significantly increased the apoptosis percentage (Fig. 5e), while the miR-422a inhibitor remarkably decreased the apoptosis percentage (Fig. 5i). The CCK-8 and MTT results indicated that miR-422a mimics decreased cell proliferation (Fig. 5f, g) and that the miR-422a inhibitor increased cell proliferation (Fig. 5j, k). In addition, overexpression of miR-422a enhanced the radiosensitization (Fig. 5h) of colorectal cancer cells, while downregulation of miR-422a exerted the opposite effects (Fig. 5l). In conclusion, these data confirmed that miR-422a suppressed cell proliferation and increased cell apoptosis and the radiosensitization of colorectal cancer by targeting MAPK1.
LINC00958 promotes the progression of colorectal cancer by targeting miR-422a
It has been revealed that lncRNAs weaken miRNA activity by competing for shared MREs, thereby regulating the expression of miRNA target genes. Therefore, we next explored whether LINC00958 could regulate the expression of MAPK1. Pearson’s correlation analysis showed that the level of LINC00958 was positively correlated with the level of MAPK1 in 63 paired colorectal cancer tissues (Fig. 6a). Knockdown of LINC00958 downregulated the expression of ERK1/2 and pERK1/2 protein and MAPK1 mRNA, downregulated the expression of Bcl-2 protein and upregulated the expression of Bax protein (Fig. 6b, c). Furthermore, si-LINC00958 decreased the luciferase activity of wild-type MAPK1 (Fig. 6d). These results implied that LINC00958 enhanced MAPK1 expression.
LINC00958 promotes MAPK1 expression and progression of colorectal cancer through miR-422a. a Pearson’s correlation showed that LINC00958 level positively correlated with MAPK1 level in 63 paired colorectal cancer tissues (R = 0.6412, p < 0.001). b The expression of MAPK1 protein (ERK1/2, pERK1/2), Bcl-2 and Bax after knockdown of LINC00958 and miR-422a in SW480 cells. c The mRNA expression of MAPK1 after knockdown of LINC00958 and miR-422a in SW480 cells. d The relative luciferase activity of wild type of MAPK1 mRNA 3’-UTR after knockdown of LINC00958 and miR-422a in SW480 cells. e The results of flow cytometry assay after knockdown of LINC00958 and miR-422a in SW480 cells. f, g The CCK-8 (f) and MTT (g) results after knockdown of LINC00958 and miR-422a in SW480 cells. h The survival fraction under different irradiation dose after knockdown of LINC00958 and miR-422a in SW480 cells. Data are reported as means ± standard deviation of three independent experiments. **p < 0.01
To test the effects of miR-422a on the roles of LINC00958 in colorectal cancer, rescue experiments were performed. The Western blot and qRT-PCR results showed that the miR-422a inhibitor relieved the suppression of si-LINC00958 on the level of ERK1/2 and pERK1/2 (MAPK1 protein) as well as the level of Bcl-2 and Bax (Fig. 6b) and MAPK1 mRNA (Fig. 6c), while the si-LINC00958 + inhibitor group was not significantly different from the si-NC group (Fig. 6b, c). Thus, the rescue experiments proved that the effects of LINC00958 on the MAPK1 pathway were purely via miR-422a. Moreover, the miR-422a inhibitor rescued the suppression of si-LINC00958 on the luciferase activity of wild-type MAPK1 (Fig. 6d). Functionally, the flow cytometry assay showed that the miR-422a inhibitor reversed the ability of si-LINC00958 to promote cell apoptosis, while the si-LINC00958 + inhibitor group was not significantly different from the si-NC group, implying that knockdown of LINC00958 increased the apoptosis percentage through miR-422a (Fig. 6e). The CCK-8 and MTT results indicated that si-LINC00958 decreased cell proliferation, while the miR-422a inhibitor relieved the ability of si-LINC00958 to inhibit cell proliferation (Fig. 6f, g). In addition, the miR-422a inhibitor increased the survival fraction under different irradiation doses downregulated by si-LINC00958, while the si-LINC00958 + inhibitor group was not significantly different from the si-NC group (Fig. 6h). Overall, these data demonstrated that si-LINC00958 suppressed cell proliferation and increased cell apoptosis and the radiosensitization of colorectal cancer by targeting miR-422a.
Discussion
To date, many lncRNAs have been revealed to play vital roles in the development and progression of various cancers. However, the functions and mechanisms of lncRNAs in the cell proliferation, apoptosis and radiosensitization of colorectal cancer have not been clearly identified. In this study, we found that LINC00958 promoted cell proliferation and suppressed cell apoptosis and radiosensitization in colorectal cancer. Mechanistically, LINC00958 served as a ceRNA to sponge miR-422a, thereby affecting its ability to suppress the expression of MAPK1.
In recent years, noncoding RNAs have attracted researchers’ attention in the exploration of biological processes. As a class of endogenous noncoding RNAs, lncRNAs are a hotspot of gene regulation, especially in the tumor research field. Abundant lncRNAs have been successfully confirmed in various cancer cell lines and different cancer types [23,24,25]. Importantly, many lncRNAs have been found to participate in various biological functions, such as cell proliferation, EMT, migration, invasion and radiosensitization. Tang et al. found that the lncRNA AATBC was upregulated in nasopharyngeal carcinoma, positively correlated with poor prognosis, and promoted cell migration and invasion [26]. The lncRNA HOTAIRM1 was reported to be downregulated in papillary thyroid cancer, significantly associated with lymph node metastasis, TNM stage and better prognosis, and suppress cell growth, invasion and migration [27]. In addition, Brownmiller et al. found a significant difference in the expression of the Y chromosome lncRNA linc-SPRY3-2/3/4 between radiation-sensitive and radiation-resistant non-small cell lung cancer cells, and confirmed that knockdown of linc-SPRY3-2/3/4 promoted cell viability and resistance to apoptosis after treatment with 8 Gy [28]. In this study, we found that LINC00958 was upregulated in colorectal cancer tissues, which was in accordance with a previous study [29]. Further analysis showed that a high level of LINC00958 was positively correlated with T stage and predicted poor overall survival and disease-free survival. Functional experiments indicated that LINC00958 remarkably enhanced cell proliferation, decreased the percentage of apoptosis and the radiosensitization of colorectal cancer cells in vitro, and promoted tumor growth in vivo. As reported in a statistical analysis of cancer data, there were no significant differences in the morbidity and mortality of colorectal cancer between males and females. Therefore, both male and female mice can be used for animal experiments. Considering that male mice generally have a stronger survival ability, only male mice were used in this animal experiment. However, to confirm whether tumor growth is sex-dependent, female mice should be studied in the future. Taken together, our results demonstrated that LINC00958 promoted cell proliferation and suppressed apoptosis and the radiosensitivity of colorectal cancer. Our data suggest that LINC00958 plays an oncogenic role in the progression of colorectal cancer and may serve as a promising diagnostic, prognostic and therapeutic marker for colorectal cancer.
Accumulating evidence shows that lncRNAs are able to act as ceRNAs to sponge miRNAs, thereby protecting target genes from being suppressed or degraded [30,31,32]. The role of ceRNAs has been widely accepted to be the main mechanism of lncRNAs in biological processes. Wei et al. demonstrated that the lncRNA HOTAIR promoted cell growth by sponging miR-1277-5p and upregulating COL5A1 expression in gastric cancer [33]. Additionally, lncRNA-SOX2OT serves as a miRNA sponge of miR-122-5p, thereby enhancing the expression of PKM2 [34]. However, the mechanisms of LINC00958 in the progression of colorectal cancer are still unidentified. RNA-FISH and nuclear-cytoplasmic fractionation assays indicated that LINC00958 was mostly located in the cytoplasm of colorectal cancer cells, presenting its possible molecular mechanism—acting as a ceRNA, which provides a direction for further study. Furthermore, bioinformatics analysis showed that LINC00958 contained a binding site of miR-422a. Pearson’s correlation showed a negative correlation between the levels of LINC00958 and miR-422a. Next, biotinylated RNA pull-down, RIP and luciferase reporter assays confirmed that LINC00958 could directly bind to miR-422a. Further rescue experiments identified that miR-422a reversed the roles of LINC00958 in colorectal cancer progression. These data suggested that LINC00958 may exert its biological function by sponging miR-422a in colorectal cancer.
MAPK1 is the main member of the mitogen-activated protein kinase family and is an important signal transmitter from the cell surface to the nucleus. The MAPK1 pathway has been confirmed to participate in many biological processes, such as cell proliferation, apoptosis, migration and invasion [35]. Radiosensitivity is one of the main factors affecting the efficacy of radiotherapy for various cancers. Soma et al. revealed that the MAPK1 pathway was involved in the radiosensitivity of multiple tumors [36]. Bioinformatics analysis showed that the MAPK1 mRNA 3’-UTR contained the binding site of miR-422a. In our study, a series of molecular experiments demonstrated that miR-422a suppressed the expression of MAPK1 by directly binding to the 3’-UTR of MAPK1 mRNA, thereby suppressing cell proliferation and increasing cell apoptosis and radiosensitization in colorectal cancer. Additionally, we found that LINC00958 promoted the expression of MAPK1. Moreover, rescue experiments demonstrated that miR-422a reversed the regulatory effect of LINC00958 on MAPK1 expression. These data revealed that LINC00958 served as a miR-422a sponge to relieve its suppression of MAPK1 expression and enhance MAPK1 gene expression.
Conclusion
We found that LINC00958 is upregulated in colorectal cancer tissues and positively correlated with clinicopathological features and poor prognosis. LINC00958 serves as a miR-422a sponge and enhances MAPK1 gene expression, thereby promoting cell proliferation and suppressing apoptosis and radiosensitivity. Our results provide insight into the progression and radiosensitivity of colorectal cancer and suggest that LINC00958 may serve as a promising diagnostic, prognostic and therapeutic marker for colorectal cancer.
Availability of data and materials
Please contact the correspondence author for the data request.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–64.
Chen C, Liu WR, Zhang B, et al. LncRNA H19 downregulation confers erlotinib resistance through upregulation of PKM2 and phosphorylation of AKT in EGFR-mutant lung cancers. Cancer Lett. 2020;486(6):58–70.
Xu J, Yang B, Wang L, et al. LncRNA BBOX1-AS1 upregulates HOXC6 expression through miR-361–3p and HuR to drive cervical cancer progression. Cell Prolif. 2020;53(7):e12823.
Wang L, Zhong Y, Yang B, et al. LINC00958 facilitates cervical cancer cell proliferation and metastasis by sponging miR-625-5p to upregulate LRRC8E expression. J Cell Biochem. 2020;121(3):2500–9.
Zhu S, Zhang J, Cui Y, et al. Long non-coding RNA HOXA11-AS upregulates Cyclin D2 to inhibit apoptosis and promote cell cycle progression in nephroblastoma by recruiting forkhead box P2. Am J Cancer Res. 2020;10(1):284–98.
Feng Y, Gao L, Cui G, Cao Y. LncRNA NEAT1 facilitates pancreatic cancer growth and metastasis through stabilizing ELF3 mRNA. Am J Cancer Res. 2020;10(1):237–48.
Zeng HF, Qiu HY, Feng FB. Long noncoding RNA LINC01133 functions as an miR-422a sponge to aggravate the tumorigenesis of human osteosarcoma. Oncol Res. 2018;26(3):335–43.
Pan K, Xie Y. LncRNA FOXC2-AS1 enhances FOXC2 mRNA stability to promote colorectal cancer progression via activation of Ca(2+)-FAK signal pathway. Cell Death Dis. 2020;11(6):434.
Tian Y, Ma R, Sun Y, et al. SP1-activated long noncoding RNA lncRNA GCMA functions as a competing endogenous RNA to promote tumor metastasis by sponging miR-124 and miR-34a in gastric cancer. Oncogene. 2020;39(25):4854–68.
Liang H, Tang Y, Zhang H, Zhang C. MiR-32-5p regulates radiosensitization, migration and invasion of colorectal cancer cells by targeting TOB1 gene. Onco Targets Ther. 2019;12(26):9651–61.
Wang M, Wang L, He X, et al. lncRNA CCAT2 promotes radiotherapy resistance for human esophageal carcinoma cells via the miR-145/p70S6K1 and p53 pathway. Int J Oncol. 2020;56(1):327–36.
Sun W, Ren S, Li R, Zhang Q, Song H. LncRNA, a novel target biomolecule, is involved in the progression of colorectal cancer. Am J Cancer Res. 2019;9(11):2515–30.
Wu H, Wei M, Jiang X, et al. lncRNA PVT1 promotes tumorigenesis of colorectal cancer by stabilizing miR-16-5p and interacting with the VEGFA/VEGFR1/AKT Axis. Mol Ther Nucleic Acids. 2020;20(12):438–50.
Hua Q, Mi B, Xu F, et al. Hypoxia-induced lncRNA-AC020978 promotes proliferation and glycolytic metabolism of non-small cell lung cancer by regulating PKM2/HIF-1α axis. Theranostics. 2020;10(11):4762–78.
Si W, Shen J, Du C, et al. A miR-20a/MAPK1/c-Myc regulatory feedback loop regulates breast carcinogenesis and chemoresistance. Cell Death Differ. 2018;25(2):406–20.
Ye Y, Guo J, Xiao P, et al. Macrophages-induced long noncoding RNA H19 up-regulation triggers and activates the miR-193b/MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma. Cancer Lett. 2020;469(3):310–22.
Gong D, Zhang J, Chen Y, et al. The m(6)A-suppressed P2RX6 activation promotes renal cancer cells migration and invasion through ATP-induced Ca(2+) influx modulating ERK1/2 phosphorylation and MMP9 signaling pathway. J Exp Clin Cancer Res. 2019;38(1):233.
Zhang K, Chen H, Zhang B, et al. Overexpression of Raf-1 and ERK1/2 in sacral chordoma and association with tumor recurrence. Int J Clin Exp Pathol. 2015;8(1):608–14.
Zuo X, Chen Z, Gao W, et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13(1):5.
Wang Z, Zhu X, Dong P, Cai J. Long noncoding RNA LINC00958 promotes the oral squamous cell carcinoma by sponging miR-185–5p/YWHAZ. Life Sci. 2020;242(23):116782.
Cheng Q, Wang L. LncRNA XIST serves as a ceRNA to regulate the expression of ASF1A, BRWD1M, and PFKFB2 in kidney transplant acute kidney injury via sponging hsa-miR-212-3p and hsa-miR-122-5p. Cell Cycle. 2020;19(3):290–9.
Zhou Z, Lin Z, He Y, et al. The long noncoding RNA D63785 regulates chemotherapy sensitivity in human gastric cancer by targeting miR-422a. Mol Ther Nucleic Acids. 2018;12(6):405–19.
Wei F, Yang L, Jiang D. Long noncoding RNA DUXAP8 contributes to the progression of hepatocellular carcinoma via regulating miR-422a/PDK2 axis. Cancer Med. 2020;9(7):2480–90.
Tang T, Yang L, Cao Y, et al. LncRNA AATBC regulates Pinin to promote metastasis in nasopharyngeal carcinoma. Mol Oncol. 2020;14(9):2251–70.
Li D, Chai L, Yu X, et al. The HOTAIRM1/miR-107/TDG axis regulates papillary thyroid cancer cell proliferation and invasion. Cell Death Dis. 2020;11(4):227.
Brownmiller T, Juric J, Ivey A, et al. Y chromosome LncRNA are involved in radiation response of male non-small cell lung cancer cells. Can Res. 2020;80(19):4046–57.
Sun Y, Liu Y, Cai Y, et al. Downregulation of LINC00958 inhibits proliferation, invasion and migration, and promotes apoptosis of colorectal cancer cells by targeting miR-3619-5p. Oncol Rep. 2020;44(4):1574–82.
Chen S, Chen JZ, Zhang JQ, et al. Silencing of long noncoding RNA LINC00958 prevents tumor initiation of pancreatic cancer by acting as a sponge of microRNA-330-5p to down-regulate PAX8. Cancer Lett. 2019;446(8):49–61.
Zhao H, Zheng GH, Li GC, et al. Long noncoding RNA LINC00958 regulates cell sensitivity to radiotherapy through RRM2 by binding to microRNA-5095 in cervical cancer. J Cell Physiol. 2019;234(12):23349–59.
Liu JQ, Deng M, Xue NN, et al. lncRNA KLF3-AS1 suppresses cell migration and invasion in ESCC By impairing miR-185-5p-targeted KLF3 inhibition. Mol Ther Nucleic Acids. 2020;20(7):231–41.
Wei Z, Chen L, Meng L, Han W, Huang LXuA. LncRNA HOTAIR promotes the growth and metastasis of gastric cancer by sponging miR-1277-5p and upregulating COL5A1. Gastric Cancer. 2020;23(6):1018–32.
Liang Y, Zhang D, Zheng T, et al. lncRNA-SOX2OT promotes hepatocellular carcinoma invasion and metastasis through miR-122-5p-mediated activation of PKM2. Oncogenesis. 2020;9(5):54.
Wei WT, Nian XX, Wang SY, et al. miR-422a inhibits cell proliferation in colorectal cancer by targeting AKT1 and MAPK1. Cancer Cell Int. 2017;17(3):91.
Ghosh S, Kumar A, Tripathi RP, Chandna S. Connexin-43 regulates p38-mediated cell migration and invasion induced selectively in tumour cells by low doses of gamma-radiation in an ERK-1/2-independent manner. Carcinogenesis. 2014;35(2):383–95.
Acknowledgements
None.
Funding
This work was supported by Henan Province 2019 overseas Research Project of Health Science and Technology Talents (HWYX2019116), Henan Medical Science and Technology Research Project (No.201303132, No.201303163, No.2018020432), Construction project of medical service capacity improvement project of medical institutions directly under Henan Province (NO.CYQ20190073).
Author information
Authors and Affiliations
Contributions
HL: Project administration; Methodology; roles/writing—original draft; software; visualization. QZ: Investigation; formal analysis; resources. ZZ: Data curation; writing—review & editing. CZ: Project administration; validation; supervision. HZ: Conceptualization; project administration; validation. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The studies involving human participants were approved by Institutional Review Boards of Henan Provence People’s Hospital. Written informed consents were acquired from all the objects.
Consent for publication
Not applicable.
Competing interests
The authors confirm that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liang, H., Zhao, Q., Zhu, Z. et al. Long noncoding RNA LINC00958 suppresses apoptosis and radiosensitivity of colorectal cancer through targeting miR-422a. Cancer Cell Int 21, 477 (2021). https://doi.org/10.1186/s12935-021-02188-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-021-02188-0