Abstract
Background
Runt related transcription factor3 (RUNX3) is considered as a tumor suppressor gene (TSG) that functions through the TGF-β dependent apoptosis. Promoter methylation of the CpG islands of RUNX3 and overexpression of enhancer of zeste homolog 2 (EZH2) has been suggested to downregulate RUNX3 in cancer.
Methods
Here, we studied the expression of RUNX3 and EZH2 in 58 esophageal tumors along with paired adjacent normal tissue. mRNA levels, protein expressions and cellular localizations of EZH2 and RUNX3 were analyzed using real-time PCR and immunohistochemistry, respectively. DNA methylation was further assessed by the methylation specific-PCR.
Results
Compared to normal tissue, a significant increase in expression of RUNX3 mRNA in 31/57 patient’s tumor tissue (p < 0.04) was observed. The expression of EZH2 was found to be upregulated compared to normal, and a significant positive correlation between EZH2 and RUNX3 expression was observed (p = 0.002). 22 of the 27 unmethylated cases at the promoter region of the RUNX3 had elevated RUNX3 protein expression (p < 0.001).
Conclusion
The data presented in this study provide new insights into the biology of RUNX3 and highlights the need to revisit our current understanding of the role of RUNX3 in cancer.
Similar content being viewed by others
Background
With increasing environmental stresses and unhealthy lifestyles, cancer has become a bane for humans with around 14.1 million new cancer cases turning up. Thousands of people every year dwell with one of the hundred types of cancer and it has been estimated that around 8.2 million people die due to cancer [1]. Esophageal cancer (CaEs), a cancer of the gastrointestinal tract has, become eighth most common cancer worldwide and, leads at sixth position in context of the deaths due to cancers [2]. 450,000 people worldwide are currently suffering from CaEs which exists majorly as esophageal squamous cell carcinoma (ESCC) and adenocarcinoma (EAC) [3, 4]. Adenocarcinoma occurs mainly in Western countries and often preceded with the GERD whereas ESCC found to be the predominant type of CaEs in Asia pacific region [5]. Treatment includes surgery, chemotherapy and radiotherapy which are either given separately or in combination with one another. However, surgery is the most opted therapy for esophageal tumor [6]. The overall prognosis of CaEs is poor with 5-year survival rates ranging between 15 and 50% [7, 8]. Therefore, there is an urgent need to identify putative targets of clinical relevance.
It is well-known that genetic mutations in the tumor suppressor and/or proto-oncogenes are responsible for initiation and progression of cancer [9,10,11]. In CaEs, many tumor suppressor genes (TSGs) have been discovered [12,13,14]. Recently, the runt-related transcription factor 3 gene (RUNX3), belonging to the runt domain family of transcription factors, has gained attention for its role in tumor progression [15]. A broader consideration revealed its conjunction with the TGF-β pathway and its upregulation which induce cell cycle arrest, apoptosis and bring down cyclin D1 expression [16,17,18,19]. Studies have indicated a tumor-suppressing role of RUNX3 [20] and complete inactivation or downregulation of RUNX3 gene has been associated with gastric cancer [17], CaEs [21, 22] pancreatic cancer [23]. RUNX3 inactivation or downregulation has shown to be dictated by hemizygous deletion [17] or mislocalization [24] or hypermethylation [25]. However, some evidence suggests that RUNX3 may have oncogenic role in cancer [26]. Accordingly, the current study attempts to investigate the hitherto unknown status of RUNX3 in Indian esophageal cancer patients.
Enhancer of zeste homolog 2 (EZH2) is known to down-regulate the expression of RUNX3. EZH2, a histone methyl transferase, is a member of polycomb group of genes (Pcg) [27]. Frequent EZH2 over-expression has been associated with cancer, however, the underlying mechanism remains unelucidated [28,29,30]. Furthermore, EZH2 is known to down-regulate the expression of RUNX3 in gastric cancers [31]. In addition, hypermethylation of RUNX3 promoter has been associated with down-regulation of RUNX3 gene expression in cancers [31, 32]. We therefore, studied the status of EZH2 and its correlation with RUNX3 expression in Indian esophageal cancer patients.
Material and methods
Selection of patient material
Tumor samples were collected from 58 esophageal cancer patients were procured from the Department of Gastrointestinal Surgery, G.B. Pant Hospital between December 2013 and March 2017. The cases selected were based on the following criteria: (i) histological proven primary ESCC with available biopsy specimens; (ii) no previous malignant disease or a second primary tumor; (iii) no previous treatment or severe complications; (iv) no chemotherapy or radiotherapy given; (v) patient belonged to the North Indian region. All the other patients who does not follow the above criterion were excluded from the study.
Recruited patient’s tumor tissue specimens were taken by surgical resection as well as from endoscopic biopsy depending on the treatment which was given to the concerned patient. Adjacent normal esophageal mucosa from same patient was used as control. The clinicopathological factors were taken into the account and every patient was followed till May 2017. Written consent letters were obtained before the tissue excision was carried out. The study was approved by the medical ethics committee of Jamia Millia Islamia as well as G.B. Pant Hospital.
Real-time PCR (qPCR)
Total RNA was isolated from ESCC tissues and the corresponding normal tissues stored in RNA later (Qiagen) using Trizol reagent (Invitrogen, Carlsbad, California, USA), and reverse transcribed into cDNA (1.2 μg) with iscript™ Reverse Transcription Reagents (Bio-Rad laboratories Inc.). PCR was performed with lightcycler® 96 SYBR Green I Master (Roche Diagnostics India Pvt Ltd) by using primers for RUNX3 (15): sense 5-GACTGTGATGGCAGG CAATGA-3 and antisense 5-CGAAGCGAAGGTCGTTGAA-3, which amplify a 101 bp product and for EZH2: sense 5-ACGTCAGATGGTGCCAGCAATA-3 and antisense 5-CCCTGACCTCTGTCTTACTTG TGGA-3, which amplify a 120 bp product. The β-Actin mRNA was also amplified as an internal control using the following primers: sense 5-AGATGTGGATCAGCAAGC AG-3 and antisense 5-GCGCAAGTTAGGTTTTGTCA-3, which amplify a 122 bp product. The real time PCR was performed on the similar lines as carried out previously [33]. Amplification cycles consisted of denaturation at 95 °C for 1 min, 35 cycles of denaturation at 94 °C for 20 s, annealing at 59 °C for 15 s, extension at 72 °C for 20 s, and a final elongation at 72 °C for 7 min. Measurements were performed in triplicates. The relative amount of mRNA was calculated as the calibrator normalized ratio using lightcycler 96 Software 1.5. The calibrator normalized ratio was measured as the following formula:
Genomic DNA extraction
High molecular weight genomic DNA was extracted from above specimens by using genomic DNA extraction kit (MDI India) as per the manufacturer's instructions. The quantity and quality of the DNA was analyzed by Nanodrop ND1000 spectrophotometer and later by running on the 1% agarose gel stained with ethidium bromide.
Methylation specific PCR (MS-PCR)
Methylation specific PCR was done as reported earlier [34]. All samples gDNA then were subjected to bisulfite conversion using the EZ DNA Methylation kit or the EZ DNA Methylation-Lightning kit (Zymo Research), by following the instruction given by the manufacturer. Bisulfite converted DNA was amplified by two different sets of primers specific to unmethylated and methylated RUNX3 sequences. The primers were designed using Methprimer tool [35]. The primer pairs for the methylated detection were in the RUNX3 promoter region: sense 5GGTTTAGTTAATGAGTTAAGGTCGC-3 and antisense 5-TCTAATAAATACGAAAACG ACCGA-3, which amplify a 193 bp product; for the unmethylated detection the primers were: sense 5-TTTAGTTAATG AGTTAAGGTTGTGA-3 and antisense 5-TCTAATAAATACAAAAACAACCAAA-3, which amplify a 190 bp product. For positive control, commercially available completely methylated and unmethylated human genomic DNA were taken whereas, double distilled water was used in place of bisulfite converted DNA for negative control. The PCR was performed in 25 µl reaction volume containing 100 ng of bisulfite converted DNA, 1.5 mM MgCl2, 200 µM of each dNTPs, 0.5 µM each of forward and reverse oligonucleotides primers, 1× PCR buffer and 1 unit of Hot Start Taq polymerase (Qiagen, Valencia, CA) hot start master mix and consisted of 35 cycles at 96 °C for 20 s; 56 °C/53 °C for 20 s; and 72 °C for 30 s after the initial denaturation step (94 °C for 5 min). A final extension was at 72 °C for 10 min. Aliquots from PCR products were visualized on 2% agarose gel containing ethidium bromide, analyzed and photographed using Gel Doc (Bio-Rad Laboratories, CA, USA) under UV illumination. As an internal quality control, each MSP was repeated and no discordant results were obtained.
Immunohistochemistry (IHC)
IHC was performed as reported earlier [36]. IHC Staining was carried on formaline fixed tissue samples. The tissue was embedded in paraffin and then cut into 4–5 µm tissue sections which were then taken on Poly-l-lysine coated slides. Xylene with differential grades of ethanol led to the deparaffinization of the tissue samples. Internal peroxidase activity was quenched by the application of 0.3% H2O2 for 30 min and subsequent 100 °C citrate buffer at pH 9 was done for Ag retrieval. Sections were blocked with TENG-T [10 mM Tris, 5 mM Ethylenediaminetetraacetic acid, 0.15 mol/l NaCl, 0.25% gelatin, 0.05% (vol/vol) Tween 20, pH 8.0] for 30 min. Slides were incubated with primary antibodies to EZH2 (1:5000) and to RUNX3 (1:4000) overnight at 4 °C in phosphate-buffered saline with 0.1% Triton and 1% bovine serum albumin. Afterwards, incubation with secondary biotinylated secondary antibody against mouse and rabbit and streptavidin horse-radish peroxidase were carried out each for 20 min. DAB was added to visualize the antibody antigen reaction, and counterstained with hematoxylin. Normal esophagus tissue was used s positive control and negative control sections for all antibodies were processed in an identical manner after omitting the primary antibody and showed no staining. staining was then interpreted by expert pathologists under light microscope at 400X magnification.
The degree of immunoreactivity of both EZH2 and RUNX3 was categorized as follows: High reactivity, more than 50% of cells showing intense immunoreactivity in their nuclei; Low reactivity, 50% of fewer cells showing intense immunoreactivity in their nuclei. The mean percentage of positive tumor cells was determined in at least five areas at high power field [37].
Statistical analysis
Data are expressed as mean ± standard deviation (SD). Statistical analysis was performed using the Statistical Package of Social Science (SPSS). The chi-square test and Fisher’s exact test were used where appropriate. The Wilcoxon signed-ranks test and Kruskal–Wallis test were drawn to assess the significance in differences at the expression levels of RUNX3/β-Actin mRNA. Spearman’s rank correlation coefficient was calculated to analyze the association between EZH2 and RUNX3 messenger RNA (mRNA) expression. p-values < 0.05 were considered as significant.
Results
Upregulated RUNX3 mRNA expression in esophageal tumors
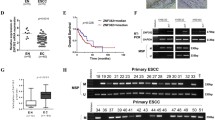
Real-time PCR was performed on cDNA from 57 CaEs tumor and adjacent normal tissues, expression of RUNX3 mRNA was found to be significantly increased in tumors (5.056 ± 5.331, relative values to β-actin expression) compared with normal tissue (5.603 ± 5.709 relative values to β-actin expression) (p < 0.04) (Fig. 1). The overall mean fold change was found to be up-regulated by 5.15 ± 10.05-fold. However, when RUNX3 upregulation was correlated with the different grades of dysphagia, no significant association was seen (p < 0.38), and the degree of association was found to be very weak. Also, no significant correlation of RUNX3 with different clinicopathological parameters was found (Table 1).
Absence of promoter DNA methylation correlated with upregulated RUNX3
In our study, we found unmethylated CpG in RUNX3 in 47.36% (27/57) samples and out of these 27 samples 81.48% (22/27) samples showed elevated expression of the RUNX3 protein in the tumor as compared to the normal tissue (Fig. 2). Whereas, MSP analysis also pointed to DNA aberrant methylation in 52.63% (30/57) of the CaEs patients and out of these only 9 samples showed upregulation. Hence, a significant correlation was seen between CpG methylation and the RUNX3 expression (p < 0.001). Also, we found that in 06 cases with methylation as well as up-regulated EZH2 protein expression RUNX3 was downregulated in tumor tissue. Whereas, 04 cases had reduced RUNX3 protein expression without corresponding methylation and 02 cases among them had up-regulated EZH2 (Table 2).
RUNX3 mRNA expression positively correlated with EZH2 mRNA and protein level
The mean fold change of expression of EZH2 mRNA was found to be > twofold up-regulated in 52 samples where expression of EZH2 was seen. The expression of EZH2 was increased in tumors (6.551 ± 1.527, relative values to B-actin expression) compared with normal tissue (6.565 ± 2.139 relative values to B-actin expression) (Fig. 3). Also, no significant association was observed between the EZH2 expression and the dysphagia grade, the degree of association also was found to be very weak (Table 3). A positive correlation was observed between the mRNA expression status of EZH2 and RUNX3. Out of 22 cases with downregulated RUNX3 protein expression 08 cases had upregulated EZH2 protein expression and 14 cases had shown downregulation (Table 4). Whereas in 23 cases of RUNX3 upregulation had EZH2 up regulation and in 06 cases downregulation of EZH2 was observed (p = 0.002).
Subcellular localization of RUNX3 and EZH2 was found to be predominantly in nucleus
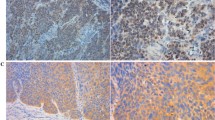
57 samples were tested for the RUNX3 and 26 samples showed low reactivity whereas 31 samples showed moderate to high reactivity. EZH2 was assessed for its expression and 21 cases were found to have low expression and in 31 cases moderate to high expression was observed (Fig. 4). These results again very well corroborated with the real time mRNA expression. All the positive cases showed nuclear expression for RUNX3 and EZH2.
Expression of RUNX3 and EZH2 as detected by IHC: a EZH2 high expression in esophageal tumor tissue. b EZH2 low expression in esophageal tumor tissue. c EZH2 expression in normal esophageal tissue. d RUNX3 high expression in tumor esophageal tissue. e Low RUNX3 expression in esophageal tumor tissue. f RUNX3 expression in normal esophageal tissue
Elevated expression of RUNX3 as revealed by oncomine database
Consistent with our findings various studies also reported overexpression of RUNX3 at mRNA level, thus pointing to a probable underlying mechanism of RUNX3 in the tumorigenesis of the esophagus. Hu dataset revealed an upregulation of RUNX3 in ESCC with a fold change of 2.661 (n = 34) Fig. 5 [38]. Another dataset of Su esophagus study and Kim esophagus study on 106 samples and 103 samples found RUNX3 overexpressing with a fold change of 1.48 and 1.29 respectively Fig. 5 [39, 40]. Some small patient dataset studies like Kimchi (n = 16) and Hao (n = 33) dataset also pointed out the upregulation of RUNX3 with a fold change of 2.337 and 7.741 respectively Fig. 5 [41, 42]. The expression of EZH2 was coincidentally also found to be overexpressing in tumor tissue in the same datasets considered earlier. Hu esophagus statistics showed EZH2 upregulated in tumor tissue by 2.09-fold Fig. 6 [38]. In Su esophagus study the fold change was 1.87 however, in Kim esophagus no change was seen in the expression Fig. 6 [39, 40]. Kimchi esophageal study and Hao esophagus study revealed EZH2 showing fold changes of 2.4 and 1.6 respectively Fig. 6 [41, 42].
Discussion
RUNX3 is known to have tumor suppressive role in gastrointestinal cancers [17, 43]. Studies have shown low level of RUNX3 expression in esophageal tumor samples and its expression has been associated with radio-resistance and poor prognosis [33, 44]. Here, we investigated the status of RUNX3 in esophageal tumors from North Indian patients. Data revealed significantly upregulated mRNA of RUNX3 as compared to the normal adjacent tissue from the same patient in ~ 55% of the samples studied. This observation was statistically significant in the distribution of the expression values of the normal and the tumor tissue in this paired study. Notably, our data suggests that RUNX3 may not be always down-regulated in esophageal cancer, as demonstrated by several studies in different cancers [20, 26]. The observation of RUNX3 up-regulation in present study highlights its plausible role in esophageal cancer. Consistent with our study, oncomine data analysis also revealed RUNX3 and EZH2 up-regulation in five studies on esophageal cancer, Oncomine™ (Compendia Bioscience, Ann Arbor, MI) was used for analysis and visualization (Figs. 5, 6).
Correlating to the observed up-regulation of mRNA expression in tumors, RUNX3 protein level was also found to be upregulated in IHC of tumor compared to normal tissues. IHC data on tumor and control tissues thus further corroborated Real Time PCR. Since several reports suggested low RUNX3 expression in GI cancers, our data, adds a new dimension to the biology of RUNX3 and suggests that RUNX3 to function in tumor suppressive manner and emphasizes the need to revisit our understanding of RUNX3 biology in GI cancers. These results again points to the debate whether RUNX3 functions as a tumor suppressor gene or as an oncogene or can act as both depending on tumor context [45, 46]. Recently it has been demonstrated that RUNX3 when associates with MYC functions as a tumor promoter whereas; acts as a tumor suppressor when interacts with p53 [47]. Various other studies have demonstrated the oncogenic role of RUNX3 [46, 48,49,50,51,52,53,54].
The methylation experiments, consistent with the previous studies, demonstrated that RUNX3 expression correlated with the methylation status of the RUNX3 promoter CpG islands. In 27 cases, we found absence of methylation at the RUNX3 promoter and out of these 27 cases, 22 cases showed up-regulation of the RUNX3; p < 0.001 (Fig. 3; Table 3). Therefore, as suggested by previous studies, RUNX3 expression can be modulated by the differential methylation status at the promoter region [55].
To explain the presence of up-regulated RUNX3 in esophageal cancer of Indian patients, we conjecture two possible explanations. Mutations in RUNX3 may render inactivated or truncated version of the RUNX3 protein. The other explanation for RUNX3 up-regulation emanates from the possibility of adaptation of cells to over-express RUNX3, as a tumor suppressor gene, to counter the induction of cancer. It would be interesting to assess the structure and activity of RUNX3 protein (as transcription factor) in tumors where it is up-regulated to establish if its active or inactive in such cases. These, along with additional functional biology studies, may provide insights into the biological relevance of RUNX3 in esophageal cancers. Whittle et al. showed that in pancreatic cancer RUNX3 upregulation was involved in the increased metastasis, hence their study showed that RUNX3 played a role of tumor suppressor as well as tumor promoter in pancreatic ductal adenocarcinoma [46]. Similar studies in esophageal cancer are needed to establish a clearer role of RUNX3 on different characteristics of cancer cells. Another possible explanation can be attributed to dietary and environmental factors of our studied population leading to disparity in RUNX3 expression.
EZH2 is frequently over-expressed in a variety of cancers and its over-expression has been implicated in the down-regulation of RUNX3 [31]. However, the results presented here suggests that EZH2 doesn’t play a role in RUNX3 down-regulation and it’s the promoter methylation that regulates the expression of RUNX3. Interestingly, our results showed up-regulation of RUNX3 coincided with the absence of methylation of RUNX3 promoter region, suggesting that methylation of CpG islands of RUNX3 promoter regulate its expression, which is in agreement with other studies [56]. The observed positive correlation between RUNX3 and EZH2 (p < 0.03) suggests the possibility of their cooperative and/or interactive role in esophageal cancer, which invites further investigation. As cancer is a complex disease with multiple genes involved, it is always pertinent to consider that possibility of cooperative and/or interactive behavior of genes and their products in the pathogenesis of cancer, for identification of viable therapeutic targets.
Conclusion
The results presented here highlights for the first time the relevance of RUNX3 and EZH2 in esophageal cancer, at least in Indian population. However, their aberrant expression in esophageal tissue biopsies also invite further investigation to be done to establish the role of RUNX3 in cancer is tumor suppressive or oncogenic.
Availability of data and materials
The datasets supporting the conclusions of this article are included within this article. Raw data are available from the corresponding author on reasonable request.
References
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–386.
Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24(14):2137–50.
Siddiqui FA, Prakasam G, Chattopadhyay S, Rehman AU, Padder RA, Ansari MA, Irshad R, Mangalhara K, Bamezai RNK, Husain M, et al. Curcumin decreases Warburg effect in cancer cells by down-regulating pyruvate kinase M2 via mTOR-HIF1α inhibition. Sci Rep. 2018;8(1):8323.
Wu J, Wu X, Liang W, Chen C, Zheng L, Adn H. Clinicopathological and prognostic significance of chemokine receptor CXCR4 overexpression in patients with esophageal cancer: a meta-analysis. Tumour Biol. 2010;35(4):3709–15.
Chung CS, Lee YC, Wang CP, Ko JY, Wang WL, Wu MS, Wang HP. Secondary prevention of esophageal squamous cell carcinoma in areas where smoking, alcohol, and betel quid chewing are prevalent. J Formos Med Assoc. 2009;109(6):408–21.
Gebski V, Burmeister B, Smithers BM, Foo K, Zalcberg J, Simes J. Survival benefits from neoadjuvant chemoradiotherapy or chemotherapy in oesophageal carcinoma: a meta-analysis. Lancet Oncol. 2007;8(3):226–34.
Rutegard M, Charonis K, Lu Y, Lagergren P, Lagergren J, Rouvelas I. Population-based esophageal cancer survival after resection without neoadjuvant therapy: an update. Surgery. 2012;152(5):903–10.
van Hagen P, Hulshof MC, van Lanschot JJ, Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, Richel DJ, Nieuwenhuijzen GA, Hospers GA, Bonenkamp JJ, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;366(22):2074–84.
Knudson AG Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68(4):820–3.
O'Driscoll M. Diseases associated with defective responses to DNA damage. Cold Spring Harb Perspect Biol. 2012;4:12.
Shih C, Weinberg RA. Isolation of a transforming sequence from a human bladder carcinoma cell line. Cell. 1982;29(1):161–9.
Kong KL, Kwong DL, Fu L, Chan TH, Chen L, Liu H, Li Y, Zhu YH, Bi J, Qin YR, et al. Characterization of a candidate tumor suppressor gene uroplakin 1A in esophageal squamous cell carcinoma. Cancer Res. 2010;70(21):8832–41.
Rasool S, Khan T, Qazi F, Ganai BA. ECRG1 and its relationship with esophageal cancer: a brief review. Onkologie. 2010;36(4):213–6.
Shibata-Kobayashi S, Yamashita H, Okuma K, Shiraishi K, Igaki H, Ohtomo K, Nakagawa K. Correlation among 16 biological factors [p53, p21(waf1), MIB-1 (Ki-67), p16(INK4A), cyclin D1, E-cadherin, Bcl-2, TNF-alpha, NF-kappaB, TGF-beta, MMP-7, COX-2, EGFR, HER2/neu, ER, and HIF-1alpha] and clinical outcomes following curative chemoradiation therapy in 10 patients with esophageal squamous cell carcinoma. Oncol Lett. 2013;5(3):903–10.
Bangsow C, Rubins N, Glusman G, Bernstein Y, Negreanu V, Goldenberg D, Lotem J, Ben-Asher E, Lancet D, Levanon D, et al. The RUNX3 gene–sequence, structure and regulated expression. Gene. 2001;279(2):221–32.
Chi XZ, Yang JO, Lee KY, Ito K, Sakakura C, Li QL, Kim HR, Cha EJ, Lee YH, Kaneda A, et al. RUNX3 suppresses gastric epithelial cell growth by inducing p21(WAF1/Cip1) expression in cooperation with transforming growth factor {beta}-activated SMAD. Mol Cell Biol. 2005;25(18):8097–107.
Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109(1):113–24.
Shiraha H, Nishina S, Yamamoto K. Loss of runt-related transcription factor 3 causes development and progression of hepatocellular carcinoma. J Cell Biochem. 2011;112(3):745–9.
Wei D, Gong W, Oh SC, Li Q, Kim WD, Wang L, Le X, Yao J, Wu TT, Huang S, et al. Loss of RUNX3 expression significantly affects the clinical outcome of gastric cancer patients and its restoration causes drastic suppression of tumor growth and metastasis. Cancer Res. 2005;65(11):4809–16.
Bae SC, Choi JK. Tumor suppressor activity of RUNX3. Oncogene. 2004;23(24):4336–400.
Chen H, Wang Z, Wang S, Zhang Z, Shi S. Effect and mechanism of RUNX3 gene on biological characteristics of human esophageal squamous cell carcinoma (ESCC). Med Oncol. 2015;32(1):357.
Long C, Yin B, Lu Q, Zhou X, Hu J, Yang Y, Yu F, Yuan Y. Promoter hypermethylation of the RUNX3 gene in esophageal squamous cell carcinoma. Cancer Invest. 2007;25(8):685–90.
Wada M, Yazumi S, Takaishi S, Hasegawa K, Sawada M, Tanaka H, Ida H, Sakakura C, Ito K, Ito Y, et al. Frequent loss of RUNX3 gene expression in human bile duct and pancreatic cancer cell lines. Oncogene. 2004;23(13):2401–7.
Ito K, Liu Q, Salto-Tellez M, Yano T, Tada K, Ida H, Huang C, Shah N, Inoue M, Rajnakova A, et al. RUNX3, a novel tumor suppressor, is frequently inactivated in gastric cancer by protein mislocalization. Cancer Res. 2005;65(17):7743–50.
Ku JL, Kang SB, Shin YK, Kang HC, Hong SH, Kim IJ, Shin JH, Han IO, Park JG. Promoter hypermethylation downregulates RUNX3 gene expression in colorectal cancer cell lines. Oncogene. 2004;23(40):6736–42.
Kumar A, Singhal M, Chopra C, Srinivasan S, Surabhi RP, Kanumuri R, Tentu S, Jagadeeshan S, Sundaram S, Ramanathan K, et al. Threonine 209 phosphorylation on RUNX3 by Pak1 is a molecular switch for its dualistic functions. Oncogene. 2016;35(37):4857–65.
Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647(1–2):21–9.
Yamaguchi H, Hung MC. Regulation and role of EZH2 in cancer. Cancer Res Treat. 2014;46(3):209–22.
Volkel P, Dupret B, Le Bourhis X, Angrand PO. Diverse involvement of EZH2 in cancer epigenetics. Am J Transl Res. 2015;7(2):175–93.
Liu F, Gu L, Cao Y, Fan X, Zhang F, Sang M. Aberrant overexpression of EZH2 and H3K27me3 serves as poor prognostic biomarker for esophageal squamous cell carcinoma patients. Biomarkers. 2016;21(1):80–90.
Fujii S, Ito K, Ito Y, Ochiai A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J Biol Chem. 2008;283(25):17324–32.
Kodach LL, Jacobs RJ, Heijmans J, Noesel CJ, Langers AM, Verspaget HW, Hommes DW, Offerhaus GJ, Brink GR, Hardwick JC. The role of EZH2 and DNA methylation in the silencing of the tumour suppressor RUNX3 in colorectal cancer. Carcinogenesis. 2010;31(9):1567–1575.
Sakakura C, Miyagawa K, Fukuda KI, Nakashima S, Yoshikawa T, Kin S, Nakase Y, Ida H, Yazumi S, Yamagishi H, et al. Frequent silencing of RUNX3 in esophageal squamous cell carcinomas is associated with radioresistance and poor prognosis. Oncogene. 2007;26(40):5927–38.
Rehman AU, Saikia S, Iqbal MA, Ahmad I, Anees A, Aravinda PS, Mishra PK, Hedau S, Saluja SS, et al. Decreased expression of MGMT in correlation with aberrant DNA methylation in esophageal cancer patients from North India. Tumour Biol. 2017. https://doi.org/10.1177/1010428317705770.
Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–31.
Siddiqui S, Chattopadhyay S, Akhtar MS, Najm MZ, Deo SV, Shukla NK, Husain SA. A study on genetic variants of Fibroblast growth factor receptor 2 (FGFR2) and the risk of breast cancer from North India. PLoS ONE. 2014;9(10):e110426.
Matsukawa Y, Semba S, Kato H, Ito A, Yanagihara K, Yokozaki H. Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci. 2006;97(6):484–91.
Hu N, Clifford RJ, Yang HH, Wang C, Goldstein AM, Ding T, Taylor PR, Lee MP. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genomics. 2010;11:576.
Kim SM, Park YY, Park ES, Cho JY, Izzo JG, Zhang D, Kim SB, Lee JH, Bhutani MS, Swisher SG, et al. Prognostic biomarkers for esophageal adenocarcinoma identified by analysis of tumor transcriptome. PLoS ONE. 2010;5(11):e15074.
Su H, Hu N, Yang HH, Wang C, Takikita M, Wang QH, Giffen C, Clifford R, Hewitt SM, Shou JZ, et al. Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin Cancer Res. 2011;17(9):2955–66.
Kimchi ET, Posner MC, Park JO, Darga TE, Kocherginsky M, Karrison T, Hart J, Smith KD, Mezhir JJ, Weichselbaum RR, et al. Progression of Barrett's metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res. 2005;65(8):3146–54.
Hao Y, Triadafilopoulos G, Sahbaie P, Young HS, Omary MB, Lowe AW. Gene expression profiling reveals stromal genes expressed in common between Barrett's esophagus and adenocarcinoma. Gastroenterology. 2006;131(3):925–33.
Saikia S, Rehman AU, Barooah P, Sarmah P, Bhattacharyya M, Deka M, Goswami B, Husain SA, Medhi S. Alteration in the expression of MGMT and RUNX3 due to non-CpG promoter methylation and their correlation with different risk factors in esophageal cancer patients. Tumour Biol. 2017;39(5):1010428317701630.
Sugiura H, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Mori Y, Ogawa R, Katada T, Harata K, Fujii Y. Decreased expression of RUNX3 is correlated with tumor progression and poor prognosis in patients with esophageal squamous cell carcinoma. Oncol Rep. 2008;19(3):713–9.
Levanon D, Bernstein Y, Negreanu V, Bone KR, Pozner A, Eilam R, Lotem J, Brenner O, Groner Y. Absence of Runx3 expression in normal gastrointestinal epithelium calls into question its tumour suppressor function. EMBO Mol Med. 2011;3(10):593–604.
Whittle MC, Izeradjene K, Rani PG, Feng L, Carlson MA, DelGiorno KE, Wood LD, Goggins M, Hruban RH, Chang AE, et al. RUNX3 controls a metastatic switch in pancreatic ductal adenocarcinoma. Cell. 2015;161(6):1345–60.
Date Y, Ito K. Oncogenic RUNX3: a link between p53 deficiency and MYC dysregulation. Mol Cells. 2020;43(2):176–81.
Damdinsuren A, Matsushita H, Ito M, Tanaka M, Jin G, Tsukamoto H, Asai S, Ando K, Miyachi H. FLT3-ITD drives Ara-C resistance in leukemic cells via the induction of RUNX3. Leuk Res. 2015;39(12):1405–13.
Lee CW, Chuang LS, Kimura S, Lai SK, Ong CW, Yan B, Salto-Tellez M, Choolani M, Ito Y. RUNX3 functions as an oncogene in ovarian cancer. Gynecol Oncol. 2011;122(2):410–7.
Lee JH, Pyon JK, Kim DW, Lee SH, Nam HS, Kang SG, Kim CH, Lee YJ, Chun JS, Cho MK. Expression of RUNX3 in skin cancers. Clin Exp Dermatol. 2011;36(7):769–74.
Barghout SH, Zepeda N, Vincent K, Azad AK, Xu Z, Yang C, Steed H, Postovit LM, Fu Y. RUNX3 contributes to carboplatin resistance in epithelial ovarian cancer cells. Gynecol Oncol. 2015;138(3):647–55.
Chen H, Crosley P, Azad AK, Gupta N, Gokul N, Xu Z, Weinfeld M, Postovit LM, Pangas SA, Hitt MM, et al. RUNX3 promotes the tumorigenic phenotype in KGN, a human granulosa cell tumor-derived cell line. Int J Mol Sci. 2019;20:14.
Nevadunsky NS, Barbieri JS, Kwong J, Merritt MA, Welch WR, Berkowitz RS, Mok SC. RUNX3 protein is overexpressed in human epithelial ovarian cancer. Gynecol Oncol. 2009;112(2):325–30.
Bledsoe KL, McGee-Lawrence ME, Camilleri ET, Wang X, Riester SM, van Wijnen AJ, Oliveira AM, Westendorf JJ. RUNX3 facilitates growth of Ewing sarcoma cells. J Cell Physiol. 2014;229(12):2049–56.
Subramaniam MM, Chan JY, Soong R, Ito K, Ito Y, Yeoh KG, Salto-Tellez M, Putti TC. RUNX3 inactivation by frequent promoter hypermethylation and protein mislocalization constitute an early event in breast cancer progression. Breast Cancer Res Treat. 2009;113(1):113–21.
Kodach LL, Jacobs RJ, Heijmans J, van Noesel CJ, Langers AM, Verspaget HW, Hommes DW, Offerhaus GJ, van den Brink GR, Hardwick JC. The role of EZH2 and DNA methylation in the silencing of the tumour suppressor RUNX3 in colorectal cancer. Carcinogenesis. 2010;31(9):1567–75.
Acknowledgements
Not applicable.
Funding
Authors thank Department of Biotechnology (NER-BPMC), Government of India, for financial support (BT/365/NE/TBP/2012).
Author information
Authors and Affiliations
Contributions
AUR: designed the study, performed the experiment, collected the data, interpreted the data and wrote the manuscript, MAI: interpreted the data, drafted and critically revised the manuscript, RSAS: helped in statistical analysis and interpretation of data, SS: helped in data acquisition, MK: helped in data acquisition, WMA: helped in analysis of clinical data, SM: helped in data acquisition, SSS: interpreted the data, provided samples, helped in analysis of clinical data, SAH: designed and guided he study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethical approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards approved by the medical ethics committee of Jamia Millia Islamia as well as G.B. Pant Hospital. Informed written consent letters were obtained before the tissue excision was carried out.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Rehman, A.U., Iqbal, M.A., Sattar, R.S.A. et al. Elevated expression of RUNX3 co-expressing with EZH2 in esophageal cancer patients from India. Cancer Cell Int 20, 445 (2020). https://doi.org/10.1186/s12935-020-01534-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-020-01534-y