Abstract
Background
Gastric cancer (GC) is one of the high-risk cancers that lacks effective methods for prognosis prediction. Therefore, we searched for immune cells related to the prognosis of GC and studied the role of related genes in GC prognosis.
Methods
In this study, we collected the mRNA data of GC from The Cancer Genome Atlas (TCGA) database and studied the immune cells that were closely related to the prognosis of GC. Spearman correlation analysis was performed to show the association between immune cell-related genes and the differentially expressed genes (DEGs) of GC. Univariate and multivariate Cox regression analyses were conducted on the immune cell-related genes with a high correlation with GC. A prognostic risk score model was constructed and the most significant feature genes were identified. Kaplan–Meier method was then used to compare the overall survival (OS) of patients with high-risk and low-risk, and receiver operating characteristic (ROC) analysis was used to assess the accuracy of the risk model. In addition, GC patients were grouped according to the median expression of the features genes, and survival analysis was further carried out.
Results
It was noted that regulatory T cells (Tregs) were significantly correlated with the prognosis of GC, and 172 genes related to Tregs were found to be closely associated with GC. An optimal prognostic risk model was constructed, and a 5-gene (including LRFN4, ADAMTS12, MCEMP1, HP and MUC15) signature-based risk score was established. Survival analysis showed significant difference in OS between low-risk and high-risk samples. ROC analysis results indicated that the risk model had a high accuracy for the prognosis prediction of samples (AUC = 0.717). The results of survival analysis on each feature gene based on expression levels were consistent with the results of multivariate Cox analysis for predicting the risk rate of the 5 genes.
Conclusion
These results proved that the 5-gene signature-based risk score could be used to predict the survival of GC patients, and these 5 genes were closely related to Tregs. These findings are of great significance for studying the role of immune cells and related immune factors in regulating the prognosis of GC.
Similar content being viewed by others
Background
Gastric cancer (GC) is a world-class malignant tumor associated with digestive system [1,2,3]. The 5-year survival rate of patients with GC is generally less than 10% [4]. Patients diagnosed with GC are usually at an advanced stage and there is still no effective treatment for advanced patients [5,6,7]. In order to find an effective treatment for GC, it is very important to study the potential molecular mechanisms.
Tumor immunotherapy is a new field of oncology research that provides a new therapeutic method for different types of tumors with metastatic characteristics [8,9,10,11]. Infiltrating immune cells seem to be the most likely target cells to improve clinical prediction and therapeutic effects [12,13,14,15]. Recent experiments of drugs acting on immunomodulatory pathways have shown that when disease-related immune pathways are modulated, they will regulate immune response of patients, which will extend the survival time of patients with a variety of cancers, including melanoma, non-small cell lung cancer (NSCLC), renal cell cancer and GC [13, 16]. Some immune factors, such as PD-L1 and tumor-infiltrating lymphocytes, have been proved to have significant immunological effects on GC [17,18,19]. However, due to technical limitations, research on immunotherapy for cancer including GC, has been limited to a few types of immune cells. Therefore, analyzing and searching for immune cells and immune cytokines associated with the prognosis of GC is the key of immunotherapy.
According to the principle of “early diagnosis and early treatment”, many methods for early diagnosis or prediction and research on molecular markers have been concerned. In recent studies, many oncogenes or suppressor genes related to GC have been reported. Ba et al. found that EIF5A2 promotes the proliferation and metastasis of GC cells, while thermo-chemotherapy can inhibit GC cell proliferation and metastasis by suppressing EIF5A2 expression [20]. Du et al. demonstrated that APC gene with high expression is a biomarker for poor prognosis in patients with stage T4 GC [21]. Although a large number of genes for prognosis and diagnosis of GC have been discovered, the genes and the multi-gene signatures used for prognosis prediction still need to be studied.
In this study, we downloaded the mRNA data of GC from the TCGA-STAD dataset to find immune cells and genes most relevant to the prognosis of GC. Based on these genes, the feature genes most related to the risk of GC were screened and a multivariate Cox regression model was constructed. The model was also used to assess the survival condition of patients with high-risk and low-risk GC. These results can provide a new reference for the prognosis of GC patients.
Methods and materials
Searching for publicly available data from TCGA database
TCGA-STAD mRNA expression data and clinical data were downloaded from the TCGA database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) by July 15, 2019, containing 407 samples (375 tumor samples, 32 normal samples).
CIBERSORT data transformation and differential analysis
The mRNA FPKM data of 407 samples were used to calculate the infiltration abundance of 22 immune cells in each sample using CIBERSORT algorithm. Then, the infiltration levels were clustered. Samples were classified into the Normal group and the Tumor group, and the infiltration levels of the 22 immune cells in two groups were evaluated. Survival analysis was performed on the immune cells with significant differences in infiltration level between the Normal group and the Tumor group. At the same time, the mRNA FPKM data were treated by differential analysis using edgeR (https://bioconductor.org/packages/release/bioc/html/edgeR.html) to obtain differentially expressed mRNAs (DEmRNAs) of GC.
Spearman correlation analysis and enrichment analysis
Spearman correlation analysis was performed on the regulatory T cells (Tregs) and DEmRNAs to screen genes with a relatively high correlation (P < 0.01, |R| > 0.15) as related genes of Tregs. Then, GO and KEGG analyses were performed on these genes.
Construction of a prognostic multivariate Cox regression model and identification of feature genes
R package “Survival” was used to conduct univariate and multivariate Cox regression analyses on previously selected genes related to Tregs, and an optimal prognostic multivariate Cox regression model was constructed to find independent prognostic factors following the program guidance using default setting. The identified prognostic factors in the risk model were validated in various aspects: (1) The patients were divided into high-risk and low-risk groups according to the median risk value of all samples. Kaplan–Meier method was used to compare the overall survival (OS) of the two groups, and the log-rank method was used for statistical analysis. (2) Receiver operating characteristic (ROC) curves were plotted using the “survivalROC” package and the area under curve (AUC) was calculated.
Results
Significant differences in the infiltration level of various immune cells between GC tumor and normal tissues
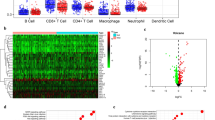
To describe our study more clearly, a flowchart of this study was presented (Fig. 1). In order to study the differences in the infiltration level of immune cells between GC tumor and normal tissues, mRNA expression data from 407 GC samples in TCGA-STAD dataset were used to generate infiltration levels of 22 major immune cells in each sample using the CIBERSORT algorithm (Fig. 2a). The data were processed by clustering analysis and the samples were divided into Normal group and Tumor group to compare the infiltration level (Fig. 2b). The results showed that there was no significant difference in infiltration level of most immune cells between the Normal group and the Tumor group. Among the cells with significant difference in infiltration level between the two groups, plasma cells, monocytes and resting mast cells showed a great decrease in the Tumor group (Fig. 2c). The infiltration levels of activated CD4 memory T cells, Tregs, M0 macrophages, M1 macrophages and M2 macrophages in Tumor group were significantly increased compared with those in the Normal group (Fig. 2c). These results indicated that the change in the infiltration level of some immune cells in tumor tissues may have a certain regulatory effect on tumorigenesis.
Significant differences in the infiltration level of various immune cells between GC and normal tissues a Infiltration abundance of 22 immune cells in each sample; b The heat map of the infiltration abundance of 22 immune cells in Tumor group (pink) and Normal group (blue). In the matrix, green to red indicates the level of infiltration gradually rises; c The difference in the infiltration level of 22 immune cells in the Normal group and the Tumor group. The blue line represents the Normal group, and the red line represents the Tumor group
Enrichment analysis of Tregs-related mRNAs
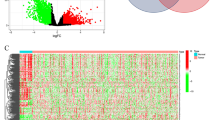
In order to study the immune cells closely related to the prognosis of GC and their related mRNAs, survival analysis was performed on 8 immune cells with significant differences in infiltration level in the Tumor group and the Normal group. The result exhibited that only the patients with a high or low infiltration level of Tregs had significant difference in OS (Fig. 3a). OS of patients with high infiltration was significantly longer than that with low infiltration, so Tregs were selected for further study. Spearman correlation analysis was performed on Tregs and DEmRNAs and 172 genes with a high correlation were screened (P < 0.01, |R| > 0.15) (Additional file 1: Table S1). GO enrichment analysis of the 172 genes revealed that these genes were enriched in a large number of immune-related biological functions, such as neutrophil activation, neutrophil degranulation and tertiary granule (Fig. 3b). The KEGG analysis also showed that most of these genes were enriched in two immune-related pathways, Rheumantoid arthritis and IL-17 signaling pathway (Fig. 3c). Therefore, Tregs may play an important role in regulating GC, and its related genes may also affect the prognosis of GC.
Enrichment analyses of Tregs-related mRNAs a Survival curves show the effect of infiltration level of Tregs on OS of patients. The blue line represents low infiltration, red line represents high infiltration; b GO enrichment analysis of Tregs-related genes; c KEGG enrichment analysis of Tregs-related genes
Establishment and evaluation of a prognostic risk model
In order to comprehensively investigate the relationship between Tregs-related genes and prognosis of GC, the 172 Tregs-related genes were used for univariate regression analysis and 38 genes with p < 0.05 were screened (Additional file 1: Table S2). These 38 genes were then used for stepwise multivariate Cox regression analysis to construct a prognostic risk model according to the program guidance. Finally, five genes (LRFN4, ADAMTS12, MCEMP1, HP, MUC15) were identified that could be used as independent prognostic factors for GC (Fig. 4). Among the 5 genes, LRFN4 was seen to have lower risk (Hazard ratio < 1) while the other 4 genes had higher risk (Hazard ratio > 1). Then, a 5-gene signature-based risk scoring formula was constructed to evaluate the survival risk of the samples according to the expression levels of the 5 genes: risk score = (–0.1634 × expression level of LRFN4) + (0.1669 × expression level of ADAMTS12) + (0.083085 × expression level of MCEMP1) + (0.078267 × expression level of HP) + (0.072812 × expression level of MUC15).
After the risk scoring formula was established, we classified each sample according to the formula to assess their survival rate. Patients were divided into high-risk and low-risk groups based on the median risk score. As exhibited in Fig. 5a, OS of the low-risk group was significantly higher than that of the high-risk group. ROC curves were also drawn to compare the sensitivity and specificity of the risk score in survival prediction (Fig. 5b). The results displayed that the AUC value was 0.717, which proved that the risk score had good sensitivity and specificity in prognosis prediction.
Kaplan–Meier survival curves of patients in high- and low-risk groups and ROC curves of the risk model a Kaplan-Meier survival curves show the effect of the 5-gene risk score on OS of patients with GC. The blue line represents low-risk group and the red line represents high-risk group; b ROC curves for assessing the sensitivity and specificity of the 5-gene risk score to predict survival risk of patients
Survival analysis of the 5 feature genes
In order to verify the accuracy of the risk assessment system, patients were respectively grouped according to the median expression of the five genes, and Kaplan–Meier curves were drawn for survival analysis. The results revealed that the survival rate of patients with high expression of ADAMTS12 and HP was significantly lower than those of patients with low expression. The survival rate of patients with high expression of LRFN4 was remarkably higher than that of patients with low expression (Fig. 6). These results were consistent with the previous conclusions.
Discussion
In this study, we used the mRNA expression data of GC to analyze and prove that there were 8 immune cells including Tregs presenting significant differences in infiltration level in GC and normal tissues. Among them, only Tregs showed significant differences in the survival rate between the high infiltration and low infiltration groups. Many previous studies reported that Tregs are associated with the development of multiple cancers including GC. Olkhanud et al. reported that tumor-induced regulatory B cells in breast cancer (BC) could transform dormant CD4 + T cells into Tregs and promote the metastasis of BC [22]. Andreas et al. studied the immunoreaction concerning p53 in head and neck cancer (HNC) and found that a certain amount of specific T cells and Tregs are clustered around p53 peptide tetramers [23]. These results suggest that Tregs are closely related to the prognosis of GC.
For purpose of studying the immune factors related to Tregs which regulate the progression of GC, we screened 172 Tregs-related genes that were closely related to GC. An optimal multivariate Cox regression model consisting of 5 genes (including LRFN4, ADAMTS12, MCEMP1, HP and MUC15) was constructed based on the 172 selected genes, and a 5-gene signature-based risk score was established. LRFN4 is regarded as a low-risk gene closely related to low-risk death and survival time prolongation. As a neuronal transmembrane protein, LRFN4 mainly plays a role in human immune system. Konakahara et al. demonstrated that LRFN4 induces monocyte/macrophage migration via actin cytoskeleton reorganization [24]. It is also the first time LRFN4 proved to be associated with GC prognosis. Therefore, the specific effects of LRFN4 on GC and other cancers need further studies. The other four genes were considered as high-risk genes in this study, and they were considered to be closely associated with high-risk death. The MCEMP1 gene is found to be associated with the prognosis of GC for the first time. It was reported that HP expression would be induced by IL-6 through activating STAT3 in HNC [25]. CHoi et al. found that MUC15 promotes the tumorigenesis of thyroid cancer and the growth of stem cells via GPCR/ERK and integrin-FAK signaling pathways [26]. However, ADAMTS12 is considered as a tumor suppressor in other cancers. Wang et al. reported that ADAMTS12 is a favorable prognostic marker in colorectal cancer, and its high expression would inhibit the occurrence of tumors [27]. In view of the differences in the roles of these feature genes in different cancers, we will conduct further studies on the specific roles of these genes in the survival of GC patients in the future.
Prognostic biomarkers refer to biomarkers related to prognosis of cancer patients, and they can be gene expression (mRNA), protein content or small molecular metabolites [28]. As prognostic markers can be independent to determine the prognosis of patients and assist the diagnosis and treatment of cancer in clinical practice, searching for prognostic markers related to cancer prognosis has become a hot research topic [29,30,31]. Many experiments have been conducted to screen and explore prognostic markers of GC, among which the most famous one is PD-L1 gene. A study has found that PD-L1 is highly expressed in cancer tissue of GC patients, which inhibits anti-tumor immune response and enables tumor cells to escape from the killing of immune cells, showing its role as the most important gene in the immune escape mechanism of cancer cells [32]. Another study has indicated that PD-L1 can be used to predict the prognosis of GC patients, but the evaluation effect is not ideal and only 43% to 63% cancer tissues of Asian cancer patients are positive for PD-L1, and the expression of PD-L1 is decreased in tumor tissues of many sufferers [33]. It is not accurate to use individual gene to judge the prognosis of cancer patients owing to the differences in race, region and organ specificity [33, 34]. In recent years, with the development of computer technology, researchers have begun to use regression models, artificial intelligence and other computational methods to screen and analyze gene expression data in cancer databases on a large scale to explore prognostic biomarkers associated with specific cancers, such as this study [35]. LRFN4, ADAMTS12, MCEMP1, HP and MUC15 are all cancer-related biomarkers, and all of them can also be used in judgment of cancer incidence in an independent manner. Compared to using PD-L1 alone to judge the prognosis of patients, this study screened prognosis-related genes based on TCGA, and the data we analyzed already involved geographical and racial differences, resulting in better applicability and accuracy in cancer diagnosis.
We established a 5-gene risk model based on the feature genes related to the survival of GC, and used it to evaluate the survival risk of the downloaded samples. The survival curves showed that the OS of patients in the high-risk group was significantly shorter than low-risk patients. This risk score further proved the important role of these feature genes in evaluating the survival of patients with GC and also demonstrated the role of Tregs-related genes in the development and prognosis of GC for the first time. Nevertheless, due to epidemiological limitations, we were unable to detect the specific correlation between this simulated risk score and the prognosis of GC. So, verification and study of the model need to be further developed.
In conclusion, we demonstrated that the 5 feature genes related to Tregs were closely associated with the prognosis of GC. This 5-gene risk score model can be used to evaluate the survival risk of patients with GC and provides a basis for studying the role of immune cells and their related factors in the prognosis of GC.
Availability of data and materials
The data used to support the findings of this study are included within the article. The data and materials in the current study are available from the corresponding author on reasonable request.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
de Martel C, Forman D, Plummer M. Gastric cancer: epidemiology and risk factors. Gastroenterol Clin North Am. 2013;42(2):219–40.
Daniyal M, Ahmad S, Ahmad M, Asif HM, Akram M, Ur Rehman S, Sultana S. Risk Factors and Epidemiology of Gastric Cancer in Pakistan. Asian Pac J Cancer Prev. 2015;16(12):4821–4.
Orditura M, Galizia G, Sforza V, Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J, Savastano B, Mabilia A, Lieto E, Ciardiello F, De Vita F. Treatment of gastric cancer. World J Gastroenterol. 2014;20(7):1635–49.
Lordick F, Kang YK, Chung HC, Salman P, Oh SC, Bodoky G, Kurteva G, Volovat C, Moiseyenko VM, Gorbunova V, Park JO, Sawaki A, Celik I, Gotte H, Melezinkova H, Moehler M. Capecitabine and cisplatin with or without cetuximab for patients with previously untreated advanced gastric cancer (EXPAND): a randomised, open-label phase 3 trial. Lancet Oncol. 2013;14(6):490–9.
Carcas LP. Gastric cancer review. J Carcinog. 2014;13:14.
Wadhwa R, Taketa T, Sudo K, Blum MA, Ajani JA. Modern oncological approaches to gastric adenocarcinoma. Gastroenterol Clin North Am. 2013;42(2):359–69.
Horn L, Spigel DR, Vokes EE, Holgado E, Ready N, Steins M, Poddubskaya E, Borghaei H, Felip E, Paz-Ares L, Pluzanski A, Reckamp KL, Burgio MA, Kohlhaeufl M, Waterhouse D, Barlesi F, Antonia S, Arrieta O, Fayette J, Crino L, Rizvi N, Reck M, Hellmann MD, Geese WJ, Li A, Blackwood-Chirchir A, Healey D, Brahmer J, Eberhardt WEE. Nivolumab versus docetaxel in previously treated patients with advanced non-small-cell lung cancer: two-year outcomes from two randomized, open-label, phase III Trials (CheckMate 017 and CheckMate 057). J Clin Oncol. 2017;35(35):3924–33.
Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O’Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016;375(19):1823–33.
Balar AV, Galsky MD, Rosenberg JE, Powles T, Petrylak DP, Bellmunt J, Loriot Y, Necchi A, Hoffman-Censits J, Perez-Gracia JL, Dawson NA, van der Heijden MS, Dreicer R, Srinivas S, Retz MM, Joseph RW, Drakaki A, Vaishampayan UN, Sridhar SS, Quinn DI, Duran I, Shaffer DR, Eigl BJ, Grivas PD, Yu EY, Li S, Kadel EE. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389(10064):67–76.
Turajlic S, Gore M, Larkin J. First report of overall survival for ipilimumab plus nivolumab from the phase III Checkmate 067 study in advanced melanoma. Ann Oncol. 2018;29(3):542–3.
Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–54.
Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, Pitot HC, Hamid O, Bhatia S, Martins R, Eaton K, Chen S, Salay TM, Alaparthy S, Grosso JF, Korman AJ, Parker SM, Agrawal S, Goldberg SM, Pardoll DM, Gupta A, Wigginton JM. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–65.
Nanda R, Chow LQ, Dees EC, Berger R, Gupta S, Geva R, Pusztai L, Pathiraja K, Aktan G, Cheng JD, Karantza V, Buisseret L. Pembrolizumab in Patients With Advanced Triple-Negative Breast Cancer: phase Ib KEYNOTE-012 Study. J Clin Oncol. 2016;34(21):2460–7.
Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, Rodig SJ, Chapuy B, Ligon AH, Zhu L, Grosso JF, Kim SY, Timmerman JM, Shipp MA, Armand P. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372(4):311–9.
Yu Y, Ma X, Zhang Y, Zhang Y, Ying J, Zhang W, Zhong Q, Zhou A, Zeng Y. Changes in expression of multiple checkpoint molecules and infiltration of tumor immune cells after neoadjuvant chemotherapy in gastric cancer. J Cancer. 2019;10(12):2754–63.
Kerr KM, Hirsch FR. Programmed Death Ligand-1 Immunohistochemistry: friend or Foe? Arch Pathol Lab Med. 2016;140(4):326–31.
Kerr KM, Nicolson MC. Non-Small Cell Lung Cancer, PD-L1, and the Pathologist. Arch Pathol Lab Med. 2016;140(3):249–54.
Wu SP, Liao RQ, Tu HY, Wang WJ, Dong ZY, Huang SM, Guo WB, Gou LY, Sun HW, Zhang Q, Xie Z, Yan LX, Su J, Yang JJ, Zhong WZ, Zhang XC, Wu YL. Stromal PD-L1-Positive Regulatory T cells and PD-1-Positive CD8-Positive T cells Define the Response of Different Subsets of Non-Small Cell Lung Cancer to PD-1/PD-L1 Blockade Immunotherapy. J Thorac Oncol. 2018;13(4):521–32.
Ba MC, Ba Z, Cui SZ, Gong YF, Chen C, Lin KP, Wu YB, Tu YN. Thermo-chemotherapy inhibits the proliferation and metastasis of gastric cancer cells via suppression of EIF5A2 expression. Onco Targets Ther. 2019;12:6275–84.
Du WB, Lin CH, Chen WB. High expression of APC is an unfavorable prognostic biomarker in T4 gastric cancer patients. World J Gastroenterol. 2019;25(31):4452–67.
Olkhanud PB, Damdinsuren B, Bodogai M, Gress RE, Sen R, Wejksza K, Malchinkhuu E, Wersto RP, Biragyn A. Tumor-evoked regulatory B cells promote breast cancer metastasis by converting resting CD4(+) T cells to T-regulatory cells. Cancer Res. 2011;71(10):3505–15.
Albers AE, Ferris RL, Kim GG, Chikamatsu K, DeLeo AB, Whiteside TL. Immune responses to p53 in patients with cancer: enrichment in tetramer + p53 peptide-specific T cells and regulatory T cells at tumor sites. Cancer Immunol Immunother. 2005;54(11):1072–81.
Konakahara S, Saitou M, Hori S, Nakane T, Murai K, Itoh R, Shinsaka A, Kohroki J, Kawakami T, Kajikawa M, Masuho Y. A neuronal transmembrane protein LRFN4 induces monocyte/macrophage migration via actin cytoskeleton reorganization. FEBS Lett. 2011;585(14):2377–84.
Li, S. C.; Lee, C. C.; Hsu, C. M.; Huang, H. B.; Su, Y. C., IL-6 induces haptoglobin expression through activating STAT3 in human head and neck cancer. J Oral Pathol Med 2019.
Choi, C.; Thi Thao Tran, N.; Van Ngu, T.; Park, S. W.; Song, M. S.; Kim, S. H.; Bae, Y. U.; Ayudthaya, P. D. N.; Munir, J.; Kim, E.; Baek, M. J.; Song, S.; Ryu, S.; Nam, K. H., Promotion of tumor progression and cancer stemness by MUC15 in thyroid cancer via the GPCR/ERK and integrin-FAK signaling pathways. Oncogenesis 2018, 7 (11), 85.
Wang D, Zhu T, Zhang FB, He C. Expression of ADAMTS12 in colorectal cancer-associated stroma prevents cancer development and is a good prognostic indicator of colorectal cancer. Dig Dis Sci. 2011;56(11):3281–7.
Oldenhuis, C. N.; Oosting, S. F.; Gietema, J. A.; de Vries, E. G., Prognostic versus predictive value of biomarkers in oncology. European journal of cancer (Oxford, England: 1990) 2008, 44 (7), 946-53.
Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, Ma P, Jiang H, Wu X, Shu Y, Xu T. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Molecular Cancer. 2020;19(1):112.
An, J. Y.; Choi, Y. Y.; Lee, J.; Hyung, W. J.; Kim, K. M.; Noh, S. H.; Choi, M. G.; Cheong, J. H., A Multi-cohort Study of the Prognostic Significance of Microsatellite Instability or Mismatch Repair Status after Recurrence of Resectable Gastric Cancer. Cancer research and treatment: official journal of Korean Cancer Association 2020.
Huang, Z.; Li, X.; Xie, T.; Gu, C.; Ni, K.; Yin, Q.; Cao, X.; Zhang, C., Elevated Expression of RIOK1 Is Correlated with Breast Cancer Hormone Receptor Status and Promotes Cancer Progression. Cancer research and treatment: official journal of Korean Cancer Association 2020.
Tang J, Yu JX, Hubbard-Lucey VM, Neftelinov ST, Hodge JP, Lin Y. Trial watch: the clinical trial landscape for PD1/PDL1 immune checkpoint inhibitors. Nat Rev Drug Discovery. 2018;17(12):854–5.
Böger C, Behrens HM, Mathiak M, Krüger S, Kalthoff H, Röcken C. PD-L1 is an independent prognostic predictor in gastric cancer of Western patients. Oncotarget. 2016;7(17):24269–83.
Vaishampayan UN, Do H, Hussain M, Schwartz K. Racial disparity in incidence patterns and outcome of kidney cancer. Urology. 2003;62(6):1012–7.
Wang, H. Y.; Chen, C. H.; Shi, S.; Chung, C. R.; Wen, Y. H.; Wu, M. H.; Lebowitz, M. S.; Zhou, J.; Lu, J. J., Improving Multi-Tumor Biomarker Health Check-up Tests with Machine Learning Algorithms. Cancers 2020, 12 (6).
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
GH contributed to the study design. NS conducted the literature search. JJ acquired the data. XC wrote the article. JJ performed data analysis and drafted. NS revised the article. GH gave the final approval of the version to be submitted. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1:
Supplementary Table 1. 172 genes with high correlation were screened (P <0.01, |R|> 0.15); Supplementary Table 2. 172 Tregs related genes were selected for univariate regression analysis and 38 genes with P < 0.05 were screened
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hu, G., Sun, N., Jiang, J. et al. Establishment of a 5-gene risk model related to regulatory T cells for predicting gastric cancer prognosis. Cancer Cell Int 20, 433 (2020). https://doi.org/10.1186/s12935-020-01502-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-020-01502-6