Abstract
Background
Endometrial carcinoma (EC) accounts for 5.8% of all cancers in Saudi females. Although most ECs are sporadic, 2–5% tend to be familial, being associated with Lynch syndrome and Cowden syndrome. In this study, we attempted to uncover the frequency, spectrum and phenotype of germline mutations in the proofreading domain of POLE and POLD1 genes in a large cohort of ECs from Middle Eastern region.
Methods
We performed Capture sequencing and Sanger sequencing to screen for proofreading domains of POLE and POLD1 genes in 432 EC cases, followed by evaluation of protein expression using immunohistochemistry. Variant interpretation was performed using PolyPhen-2, MutationAssessor, SIFT, CADD and Mutation Taster.
Results
In our cohort, four mutations (0.93%) were identified in 432 EC cases, two each in POLE and POLD1 proofreading domains. Furthermore, low expression of POLE and POLD1 was noted in 41.1% (170/1414) and 59.9% (251/419) of cases, respectively. Both the cases harboring POLE mutation showed high nuclear expression of POLE protein, whereas, of the two POLD1 mutant cases, one case showed high expression and another case showed low expression of POLD1 protein.
Conclusions
Our study shows that germline mutations in POLE and POLD1 proofreading region are a rare cause of EC in Middle Eastern population. However, it is still feasible to screen multiple cancer related genes in EC patients from Middle Eastern region using multigene panels including POLE and POLD1.
Similar content being viewed by others
Background
Endometrial carcinoma (EC) is the second most common gynecologic cancer Worldwide, with its annual incidence projected to increase [1]. In Saudi Arabia, EC is the fourth most common malignancy among women, accounting for 5.8% of all cancers in females [2]. Numerous genetic mutations have been discovered in the past few years leading to a better understanding of hereditary syndromes associated with malignancies of the female genital tract [3]. Although majority of ECs are sporadic, 2–5% tend to be familial [4]. Familial EC has been linked to germline mutations in the mismatch repair genes associated with Lynch Syndrome (LS), or to germline mutations in PTEN associated with Cowden Syndrome [5, 6]. A recent study has shown that germline missense mutations of POLE and POLD1 genes lead to development of polymerase proofreading-associated polyposis, which is similar to LS with regards to tumor spectrum, including an increased risk of ECs [7].
POLE and POLD1 are related B family polymerases. They form the major catalytic and proofreading subunits of the DNA polymerase Epsilon (Polε) and DNA polymerase Delta (Polδ) enzyme complexes [8]. Both Polε and Polδ are heterotetramers with Polymerase ε involved in replication of leading strand of the replication fork [9], whereas DNA polymerase δ functions in synthesizing the lagging strand [10]. Both polymerases δ and ε are responsible for carrying out high fidelity DNA synthesis and mutation affecting the proofreading activity of these genes can lead to genome instability, and subsequent increased risk of developing cancer [11].
POLE mutations constitute a specific molecular subgroup of EC, and have both prognostic and therapeutic implications for the patient [12]. The Cancer Genome Atlas (TCGA) characterized 373 cases of EC, based on their integrated genomic, transcriptomic, and proteomic data, into four molecular subgroups. Tumors with POLE mutations were identified as one of the subgroups and represented an ultra-mutated tumor phenotype [12]. Somatic mutations of POLE gene have been reported in 6–10% of ECs and 1–2% of colorectal cancers [12,13,14,15]. Few cases of lung, breast, stomach, pancreatic, brain and ovarian tumors have also been shown to harbor these mutations [16, 17]. Although rare, germline POLE mutations have been reported in 0.25–4% of ECs [18,19,20].
Currently, there is no known prognostic significance associated with POLD1 mutation. Instead, emphasis is placed on identification of POLD1 germline mutations due to the potential risk of developing secondary tumors in a hereditary syndromic manner [21].
With the advent of individualized therapy, greater emphasis is placed on identifying specific genetic alterations and molecular subtypes of EC. In this study, we report the frequency, spectrum and phenotype of germline mutations in the proofreading domains of POLE and POLD1 genes in a large cohort of ECs from Middle Eastern region. It may contribute to a better understanding of the molecular mechanisms underlying EC and could also have important preventive and/or therapeutic implications in Middle Eastern population.
Materials and methods
Sample selection
Archival samples from 432 EC patients diagnosed between 1990 and 2016 at King Faisal Specialist Hospital and Research Center (Riyadh, Saudi Arabia) were included in the study. Detailed clinicopathological data were noted from case records and have been summarized in Table 1. All samples were obtained from patients with approval from Institutional Review Board of the hospital. For the study, waiver of consent was obtained for archived paraffin tissue blocks from Research Advisory Council (RAC) under project RAC# 2180 001.
DNA extraction
DNAs were isolated from formalin-fixed, paraffin-embedded (FFPE) EC non-tumor tissues using Gentra DNA isolation kit (Gentra, Minneapolis, MN, USA) following the manufacturer’s recommendations as described previously [22].
Targeted capture sequencing of germline mutations in proofreading domain of POLE and POLD1 genes
The capture sequencing was performed on 53 EC cases as described previously [23]. The DNA samples with A260/A280 ratio between 1.8 and 2.0 were processed for library construction. The sequencing library was prepared by random fragmentation of the DNA, followed by 5′ and 3′ adapter ligation. Adapter-ligated fragments were then PCR amplified and gel purified. Clusters were generated by loading the library into a flow cell where fragments were captured on a lawn of surface-bound oligos complementary to the library adapters. Each fragment was then amplified into distinct, clonal clusters through bridge amplification. Raw data was generated utilizing HCS (HiSeq control software v3.3) and RTA (real-time analysis. v2.5.2).
The BCL (base calls) generated by Illumina Hiseq 4000 were converted into FASTQ files by bcl2fastq (v2.16). The sequence reads in fastq format from each sample were aligned to the reference human genome (GRCh37/hg19) using burrows-wheeler aligner (BWA) [24]. BAM file generation, PCR duplicates and local realignment was performed using Picard-tools and genome analysis toolkit (GATK) [25].
The variant calling was performed by GATK, subsequently the variants were annotated by ANNOVAR [26], with dbSNP138, 1000 Genomes, ESP6500, Exome Aggregation Consortium (ExAC), Clinvar and other genome databases.
Polymerase Chain Reaction (PCR) and Sanger Sequencing for detection of Germline Mutations in Proofreading Domain of POLE and POLD1 genes
Direct sequencing of the entire coding/splicing region of proofreading domain of POLE and POLD1 genes were performed on 379 samples. In addition, detected mutations by Capture sequencing were further confirmed by Sanger sequencing in 53 cases. Primer 3 software was used to design the primers for all coding exons and their flanking intronic sequences of proofreading domain of POLE and POLD1 genes (available upon request). PCR was performed in a total volume of 25 μL using 20 ng of genomic DNA, 2.5 μL 10× Taq buffer, 2.3 mM MgCl2, 0.2 mM dNTPs, 1 unit Taq polymerase (all reagents were from Qiagen Inc), 0.2 μM of each primer, and water. The efficiency and quality of the amplified PCR products were confirmed by running the PCR products on a 2% agarose gel.
For Sanger sequencing, the PCR products were subsequently subjected to direct sequencing with BigDye terminator V 3.1 cycle sequencing reagents and analyzed on an ABI 3730XL DNA analyzer (Applied Biosystems, Foster City, CA). Reference sequences were downloaded from NCBI GenBank. Sequencing results were compared with the reference sequences by Mutation Surveyor V4.04 (Soft Genetics, LLC, State College, PA).
Assessment of Pathogenicity of Variants
ACMG/AMP 2015 guideline was utilized first for interpretation of sequence variants [27]. All the uncertain significant variants interpreted by ACMG/AMP 2015 guideline were further analyzed using five in silico pathogenicity prediction tools: PolyPhen-2 [28], MutationAssessor [29], SIFT [30], CADD [31] and Mutation Taster [32]. The variants predicted as damaging or possibly damaging by three or more in silico prediction tools were considered as pathogenic mutations.
Tissue microarray construction and Immunohistochemistry
All samples were analyzed in a tissue microarray (TMA) format. TMA construction was performed as described earlier [33]. Briefly, tissue cylinders with a diameter of 0.6 mm were punched from representative tumor regions of each donor tissue block and brought into recipient paraffin block using a modified semiautomatic robotic precision instrument (Beecher Instruments, Woodland, WI). Two cores of EC were arrayed from each case.
Standard protocol was followed for immunohistochemistry (IHC) staining. For antigen retrieval, Dako (Dako Denmark A/S, Glostrup, Denmark) Target Retrieval Solution pH 9.0 (Catalog number S2367) was used, and the slides were placed in Pascal pressure cooker for 8 min at 120 °C. The slides were incubated with primary antibodies against POLE (ab-134941, Abcam, Cambridge, UK) and POLD1 (ab-186407, Abcam, Cambridge, UK) at a dilution of 1:1000 (pH 9.0). The Dako Envision Plus System kit was used as the secondary detection system with 3, 30-diaminobenzidine as chromogen. All slides were counterstained with hematoxylin, dehydrated, cleared and mounted. Negative controls included omission of the primary antibody. Normal tissues of different organ systems were also included in the TMA to serve as control. Only fresh cut slides were stained simultaneously to minimize the influence of slide aging and maximize reproducibility of the experiment.
Each TMA spot was assigned an intensity score from 0 to 3 (I0–I3) corresponding to no, weak, moderate and strong staining, and the proportion of tumor staining for that intensity was recorded as 5% increments from a range of 0–100 (P0–P3). A final H score (range 0–300) was obtained by adding the products of scores obtained for each intensity and proportion of area stained (H score = I1XP1 + I2XP2 + I3XP3). Using X-tile version 3.6.1 [34], we defined the optimal cutoff point for POLE and POLD1 expression as H = 90 and H = 175, respectively. Based on H scores, EC cases were classified into two subgroups: those below the cutoff score were defined as low expression and those above the cutoff score were defined as over expression.
Staining and evaluation of mismatch repair proteins (MLH1, MSH2, MSH6 and PMS2) was performed as described previously [35].
Statistical analysis
Contingency table analysis and Chi square tests were used to study the relationship between clinico-pathological variables and protein expression or mutation. Overall Survival curves were generated using the Kaplan–Meier method, with significance evaluated using the Mantel–Cox log-rank test. The limit of significance for all analyses was defined as p value of < 0.05; two-sided tests were used in these calculations. The JMP11.0 (SAS Institute, Inc., Cary, NC) software package was used for data analyses.
Results
Sample characteristics
A total of 432 EC cases were analyzed. Median age of the study cohort was 59 years. Tumors were predominantly of type I EC (88.1%) with an almost equal distribution among the three grades. Majority of the cases were Stage I tumors (64.3%) (Table 1).
Identification of Germline Mutations in Proofreading Domain of POLE and POLD1 genes
Among 53 EC cases sequenced using Capture sequencing, no mutations were identified in the proofreading domains of POLE and POLD1 genes. Among 379 EC cases analyzed by Sanger sequencing, four variants (1%, 4/379) were detected, two in POLE (0.53%) and two in POLD1 (0.53%) proofreading domain and interpreted as of uncertain significance by ACMG/AMP 2015 guideline. Further analysis utilizing in silico pathogenicity prediction tools showed that all four were pathogenic mutations; c.1403A > G;p.468Y > C and c.940T > G;p.314S > A in POLE gene and c.1120G > A;p.374E > K and c.1231C > T;p.411Q > X in POLD1 gene. Altogether, four variants (0.93%) were predicted to be pathogenic in 432 EC cases (Table 2).
The POLE p.314S > A is completely conserved and is also found in population database at a very low frequency of 0.00008 (ExAC). Another proofreading domain mutation p.468Y > C in POLE gene is also highly conserved and is completely absent in the population database of ExAC (Table 2).
The POLD1 proofreading domain mutation, p.374E > K, was detected in a patient with early onset of EC. In addition, the p.411Q > X is partially conserved and p.374E > K is completely conserved in 6 species. Furthermore, these mutations are totally absent in the database of ExAC and were predicted as pathogenic by all five in silico prediction tools (Table 2).
Both cases harboring POLE mutations were older than 60 years, with one of them being serous (grade 3) and the other being endometrioid (grade 1) EC. The patient with POLD1 p.374E > K mutation had grade 1 endometrioid EC. Another patient with POLD1 p.411Q > X mutation was older than 60 years with grade 3 serous EC (Table 2). All the four cases harboring POLE/POLD1 mutations were mismatch repair proficient as assessed by IHC.
POLE and POLD1 expression in EC and their association with clinico-pathological features
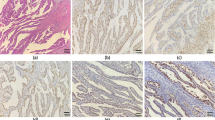
We next evaluated the expression of POLE and POLD1 by immunohistochemistry in 432 EC cases using tissue microarray. POLE immunohistochemical expression was interpretable in 414 cases. Low expression of POLE was noted in 41.1% (170/414) of cases and showed a significant association with grade 2 tumors (p = 0.0308). Both the cases harboring POLE mutation showed high nuclear expression of POLE protein. There was no significant association between POLE expression and microsatellite instability status (Table 3, Fig. 1a, b). POLD1 expression was interpretable in 419 cases. Low expression of POLD1 was noted in 59.9% (251/419) of cases and was significantly associated with grade 1 tumors (p = 0.0024) and a trend was noted with Type I EC (p = 0.0728). Of the two cases with POLD1 mutation, one case showed high expression and another case showed low expression of POLD1 protein (Table 4, Fig. 1c, d).
Tissue microarray based immunohistochemistry analysis of POLE and POLD1 in Endometrial carcinoma (EC) patients. EC TMA spots showing overexpression of POLE (a) and POLD1 (c). In contrast, another set of TMA spots showing reduced expression of POLE (b) and POLD1 (d). 20 X/0.70 objective on an Olympus BX 51 microscope (Olympus America Inc, Center Valley, PA, USA) with the inset showing a 40X 0.85 aperture magnified view of the same TMA spot
Discussion
Pathogenic mutations involving the proofreading domains of POLE and POLD1 are widely known to be associated with colorectal polyposis and cancer [14, 36]. However, their role in EC is less well established. Here, we screened the proofreading domain of POLE and POLD1 to detect causative variants in 432 unselected EC cases from the Middle Eastern region. We found two heterozygous mutations each in POLE (0.46%; 2/432) and POLD1 genes (0.46%; 2/432). To the best of our knowledge, this is the first study to determine the frequency of germline POLE and POLD1 mutations in EC from the Middle Eastern region. McConechy et al. [18] and Church et al. [19] have also reported a similar frequency for POLE and POLD1 germline mutations in EC. A study from South East Asia reported a frequency of 4.3% each for POLE and POLD1 germline mutations. However, the study was performed on only 47 selected cases of grade 3 endometrioid endometrial carcinomas [20] (Table 5). Other studies have reported the prevalence of POLE and POLD1 mutations at the somatic level, varying between 6.1 and 9.7% [12, 13, 37,38,39].
Previous studies have shown that POLE proofreading-mutant cancers are a molecularly distinct group of tumors with a striking mutation burden and distinctive mutation signature [12, 19]. We have shown that POLE p.314S > A and p.468Y > C mutations are completely conserved between 6 species and found in population database at a very low frequency or absent respectively. Interestingly, this mutation (POLE p.314S > A) was predicted as colorectal carcinoma predisposing mutation in another study by our group (data unpublished). One of the POLE mutant cases was a grade 3 serous EC and the other was grade 1 endometrioid EC. Church et al. [19] also reported a single germline POLE mutation in grade 3 endometrioid EC. However, family history information of these mutation carriers are not available due to Middle Eastern conservative culture [40].
Two mutations in POLD1, p.374 E > K and p.411 Q > X, were also detected in patients with grade 1 endometrioid and grade 3 serous EC, respectively. These variants were not found in ExAC database. These mutations were partially conserved and predicted as pathogenic mutation by at least three in silico prediction tools. The POLD1 p.411 Q > X mutation caused truncation of the protein in proofreading domain which would have adverse effect on the exonuclease activity of the gene, rendering this mutation highly pathogenic in nature. Consistent with previous reports, all the four cases harboring POLE or POLD1 mutations in our cohort were MSS tumors [12, 19, 41].
However, three out of four (75%) germline mutations identified were completely novel and weren’t reported previously in public database of ClinVar or other studies [42, 43], which could reflect the uniqueness of Saudi population (isolation, tribal origin and high consanguinity). The Gene Ontology (GO) analysis revealed POLE and POLD1 genes affect important biological processes including DNA replication proofreading and base-excision repair (Additional file 1: Table S1). It has been studied previously that loss of proofreading activity of replicative DNA polymerases and base-excision repair is responsible for various sporadic and hereditary cancers [44].
Several studies have reported favorable outcomes for women with POLE-mutated EC. This favorable prognosis has been attributed to the high number of mutations in tumors, expression of neoantigens, as well as an increase in patient immune responses [45]. Consistent with previous reports [18, 46], we observed no EC-related deaths or evidence of recurrent tumors in both patients with POLE-mutant cancers. However, we do acknowledge that the small number of tumors with POLE mutations limits our power, and therefore our results do not meet traditional levels of statistical significance. Despite this, our data contributes to the existing literature.
In this study, we showed that proofreading domain mutations in POLE and POLD1 genes were not associated with protein expression of POLE and POLD1. This result could be partly explained by the fact that somatic proofreading domain mutations were not assessed. Interestingly, Campbell et al. [47] previously reported a low number of truncated mutations in proofreading domain as compared to the region outside of proofreading domain, and one-third of truncated POLE and POLD1 mutations did not cause high tumor mutation burden. In addition, Elsayed et al. [48] also reported that the two POLE variant carriers in their cohort demonstrated positive POLE protein expression, which emphasizes the fact that POLE IHC does not have predictive value for effect of mutation. All these results indicated that POLE and POLD1 IHC analysis might not be suitable to select the patients for immunotherapy using immune checkpoint inhibitors.
Conclusions
Our study shows a low frequency of germline mutations in POLE and POLD1 proofreading domains in Middle Eastern EC patients. Although rare, screening for these mutations in individuals with high risk of developing EC might be clinically valuable. Since next generation sequencing technology offers significant benefits compared to single gene testing by reducing costs, time and increasing the sensitivity, it is feasible to screen multiple cancer related genes in EC patients using multigene panels including POLE and POLD1.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Abbreviations
- EC:
-
endometrial carcinoma
- LS:
-
lynch syndrome
- POLE:
-
polymerase epsilon
- POLD1:
-
polymerase delta 1
- TCGA:
-
The Cancer Genome Atlas
- RAC:
-
Research Advisory Council
- FFPE:
-
Formalin-Fixed Paraffin Embedded
- PCR:
-
polymerase chain reaction
- TMA:
-
tissue microarray
- IHC:
-
immunohistochemistry
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. 2018;68(6):394–424.
Al-Shahrani Z, Al-Rawaji A, Al-Madouj A, Hayder M, Al-Zahrani A, Al-Mutlaq HJRSHC, Saudi Cancer Registry: Cancer Incidence Report, Saudi Arabia 2014. 2014.
Wong A, Ngeow J. Hereditary syndromes manifesting as endometrial carcinoma: how can pathological features aid risk assessment? BioMed Res Int. 2015;2015:219012.
O’Hara AJ, Bell DW. The genomics and genetics of endometrial cancer. Adv Genom Genet. 2012;2012(2):33.
Meyer LA, Broaddus RR, Lu KH. Endometrial cancer and Lynch syndrome: clinical and pathologic considerations. Cancer Control. 2009;16(1):14–22.
Pilarski R, Eng C. Will the real Cowden syndrome please stand up (again)? Expanding mutational and clinical spectra of the PTEN hamartoma tumour syndrome. J Med Genet. 2004;41(5):323–6.
Tomlinson I. An update on the molecular pathology of the intestinal polyposis syndromes. Diagn Histopathol. 2015;21(4):147–51.
McElhinny SAN, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30(2):137–44.
Burgers PM, Gordenin D, Kunkel TA. Who is leading the replication fork, Pol ε or Pol δ? Mol Cell. 2016;61(4):492–3.
Larrea AA, Lujan SA, McElhinny SAN, Mieczkowski PA, Resnick MA, Gordenin DA, Kunkel TA. Genome-wide model for the normal eukaryotic DNA replication fork. Proc Natl Acad Sci. 2010;107(41):17674–9.
Loeb LA, Monnat RJ Jr. DNA polymerases and human disease. Nat Rev Genet. 2008;9(8):594.
Levine DA, Network CGAR. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67.
Church DN, Stelloo E, Nout RA, Valtcheva N, Depreeuw J, Haar N, Noske A, Amant F, Tomlinson IP, Wild PJ. Prognostic significance of POLE proofreading mutations in endometrial cancer. J Natl Cancer Inst. 2015;107(1):402.
Palles C, Cazier J-B, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45(2):136.
Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330.
Shinbrot E, Henninger EE, Weinhold N, Covington KR, Göksenin AY, Schultz N, Chao H, Doddapaneni H, Muzny DM, Gibbs RA. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014;24(11):1740–50.
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415.
McConechy MK, Talhouk A, Leung S, Chiu D, Yang W, Senz J, Reha-Krantz LJ, Lee C-H, Huntsman DG, Gilks CB. Endometrial carcinomas with POLE exonuclease domain mutations have a favorable prognosis. Clin Cancer Res. 2016;22(12):2865–73.
Church DN, Briggs SE, Palles C, Domingo E, Kearsey SJ, Grimes JM, Gorman M, Martin L, Howarth KM, Hodgson SV. DNA polymerase ɛ and δ exonuclease domain mutations in endometrial cancer. Hum Mol Genet. 2013;22(14):2820–8.
Wong A, Kuick CH, Wong WL, Tham JM, Mansor S, Loh E, Jain S, Vikas NN, Tan SH, Chan SH. Mutation spectrum of POLE and POLD1 mutations in South East Asian women presenting with grade 3 endometrioid endometrial carcinomas. Gynecol Oncol. 2016;141(1):113–20.
Bellido F, Pineda M, Aiza G, Valdés-Mas R, Navarro M, Puente DA, Pons T, González S, Iglesias S, Darder E. POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: review of reported cases and recommendations for genetic testing and surveillance. Genet Med. 2016;18(4):325.
Abubaker J, Jehan Z, Bavi P, Sultana M, Al-Harbi S, Ibrahim M, Al-Nuaim A, Ahmed M, Amin T, Al-Fehaily M. Clinicopathological analysis of papillary thyroid cancer with PIK3CA alterations in a Middle Eastern population. J Clin Endocrinol Metab. 2008;93(2):611–8.
Siraj AK, Masoodi T, Bu R, Parvathareddy SK, Al-Badawi IA, Al-Sanea N, Ashari LH, Abduljabbar A, Alhomoud S, Al-Sobhi SS. Expanding the spectrum of germline variants in cancer. Hum Genet. 2017;136(11–12):1431–44.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248.
Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118.
Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc. 2016;11(1):1.
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2018;47(D1):D886–94.
Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361.
Siraj A, Bavi P, Abubaker J, Jehan Z, Sultana M, Al-Dayel F, Al-Nuaim A, Alzahrani A, Ahmed M, Al-Sanea O. Genome-wide expression analysis of Middle Eastern papillary thyroid cancer reveals c-MET as a novel target for cancer therapy. J Pathol. 2007;213(2):190–9.
Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–9.
Siraj AK, Prabhakaran S, Bavi P, Bu R, Beg S, Hazmi MA, Al-Rasheed M, Al-Assiri M, Sairafi R, Al-Dayel F. Prevalence of Lynch syndrome in a Middle Eastern population with colorectal cancer. Cancer. 2015;121(11):1762–71.
Valle L, Hernández-Illán E, Bellido F, Aiza G, Castillejo A, Castillejo M-I, Navarro M, Seguí N, Vargas G, Guarinos C. New insights into POLE and POLD1 germline mutations in familial colorectal cancer and polyposis. Hum Mol Genet. 2014;23(13):3506–12.
Billingsley CC, Cohn DE, Mutch DG, Hade EM, Goodfellow PJ. Prognostic significance of POLE exonuclease domain mutations in high-grade endometrioid endometrial cancer on survival and recurrence: a subanalysis. Int J Gynecol Cancer. 2016;26(5):933–8.
Haruma T, Nagasaka T, Nakamura K, Haraga J, Nyuya A, Nishida T, Goel A, Masuyama H, Hiramatsu Y. Clinical impact of endometrial cancer stratified by genetic mutational profiles, POLE mutation, and microsatellite instability. PLoS ONE. 2018;13(4):e0195655.
Imboden S, Nastic D, Ghaderi M, Rydberg F, Rau TT, Mueller MD, Epstein E, Carlson JW. Phenotype of POLE-mutated endometrial cancer. PLoS ONE. 2019;14(3):e0214318.
Khalil RB. Attitudes, beliefs and perceptions regarding truth disclosure of cancer-related information in the Middle East: a review. Palliat Support Care. 2013;11(1):69–78.
Briggs S, Tomlinson I. Germline and somatic polymerase ϵ and δ mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol. 2013;230(2):148–53.
Esteban-Jurado C, Giménez-Zaragoza D, Muñoz J, Franch-Expósito S, Álvarez-Barona M, Ocaña T, Cuatrecasas M, Carballal S, López-Cerón M, Marti-Solano M. POLE and POLD1 screening in 155 patients with multiple polyps and early-onset colorectal cancer. Oncotarget. 2017;8(16):26732.
Pätzold L, Bērziņa D, Daneberga Z, Gardovskis J, Miklaševičs E. Detection of allelic variants of the POLE and POLD1 genes in colorectal cancer patients. Balkan J Med Genet. 2017;20(2):83–7.
Kane DP, Shcherbakova PV. A common cancer-associated DNA polymerase ε mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res. 2014;74(7):1895–901.
van Gool IC, Eggink FA, Freeman-Mills L, Stelloo E, Marchi E, de Bruyn M, Palles C, Nout RA, de Kroon CD, Osse EM. POLE proofreading mutations elicit an antitumor immune response in endometrial cancer. Clin Cancer Res. 2015;21(14):3347–55.
Van Gool IC, Rayner E, Osse EM, Nout RA, Creutzberg CL, Tomlinson IP, Church DN, Smit VT, de Wind N, Bosse T. Adjuvant treatment for POLE proofreading domain-mutant cancers: sensitivity to radiotherapy, chemotherapy, and nucleoside analogues. Clin Cancer Res. 2018;24(13):3197–203.
Campbell BB, Light N, Fabrizio D, Zatzman M, Fuligni F, de Borja R, Davidson S, Edwards M, Elvin JA, Hodel KP. Comprehensive analysis of hypermutation in human cancer. Cell. 2017;171(5):1042–56.
Elsayed FA, Kets CM, Ruano D, Van Den Akker B, Mensenkamp AR, Schrumpf M, Nielsen M, Wijnen JT, Tops CM, Ligtenberg MJ. Germline variants in POLE are associated with early onset mismatch repair deficient colorectal cancer. Eur J Hum Genet. 2015;23(8):1080.
Acknowledgements
We thank Zeeshan Qadri, Saud Azam, Valorie Balde, Padmanaban Annaiyappanaidu, Felisa De Vera and Mary Beleno Galvez for their technical assistance.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
AKS and SKP: Designed, performed experiments and wrote the manuscript. SKP: Prepared the TMA and conducted all the immunohistochemistry experiments and scoring of IHC spots. RB, KI, RMC and LG: Performed experiments. SS, TM, IAB and FA: Collected and analyzed all the clinical samples and data. KSA: Made substantial contributions to conception, design and acquisition of data along with analysis and interpretation of data; Prepared and wrote the manuscript. KSA gave the final approval for the submission of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All archival samples were obtained with approval from Institutional Review Board of the hospital. For the study, waiver of consent was obtained for archived paraffin tissue blocks from Research Advisory Council (RAC) under Project RAC# 2180 001.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Functional analysis of POLE and POLD1 genes using Gene Ontology (GO).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Siraj, A.K., Parvathareddy, S.K., Bu, R. et al. Germline POLE and POLD1 proofreading domain mutations in endometrial carcinoma from Middle Eastern region. Cancer Cell Int 19, 334 (2019). https://doi.org/10.1186/s12935-019-1058-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-019-1058-9