Abstract
Background/aims
ErbB3 is an oncogene which has proliferation and metastasis promotion effects by several signaling pathways. However, the individual expression difference regulated by miRNA was almost still unknown. We focused on the miRNAs associated SNPs in the 3′-UTR of ErbB3 to investigate the further relationship of the SNPs with miRNAs among Chinese gastric cancer (GC) patients.
Methods
We performed case–control study including 851 GC patients and 799 cancer-free controls. Genotyping, real-time PCR assay, cell transfection, the dual luciferase reporter assay, western-blot, cell proliferation and trans-well based cell invasion assay were used to investigate the effects of the SNP on ErbB3 expression. Moreover, a 5-years-overall survival and relapse free survival were investigated between different genotypes.
Results
We found that patients suffering from Helicobacter pylori (Hp.) infection indicated to be the susceptible population by comparing with controls. Besides, SNP rs3202538 (G/T) in ErbB3 3′-UTR was involved in the occurrence of GC by acting as tumor risk factors. SNP rs3202538 (G/T) could be regulated by both miR-204 and miR-211 which caused an upregulation of ErbB3 in patients. Furthermore, the carriers of T genotype was related to the significantly high expression of ErbB3, and to big tumor size, poor differentiation as well as the high probability of metastasis. Both miR-211 and miR-204 can significantly decrease cell proliferation, metastasis as well as downstream AKT activation through G but not T allele of ErbB3 3′UTR. Moreover, the SNP of G/T was associated with shorter survival of post-surgery GC patients with 5 years of follow up study.
Conclusion
In conclusion, our findings have shown that the SNP rs3202538 (G/T) in ErbB3 3′-UTR acted as promotion factors in the GC development through disrupting the regulatory role of miR-204 and miR-211 in ErbB3 expression.
Similar content being viewed by others
Introduction
Globally, gastric cancer (GC) is the fifth leading cause of cancer and the third leading cause of death from cancer making up 7% of cases and 9% of deaths. In 2012 GC occurred in 950,000 people and caused 723,000 deaths [1]. GC occurs most commonly in East Asia and Eastern Europe and it occurs twice as often in males as in females. The most common cause is infection by the bacterium Helicobacter pylori (Hp), which accounts for more than 60% of cases. The current diagnostic system was proved to be relatively poor in early-stage diagnosis GC, and accumulating evidence revealed the great potential of microRNAs (miRNAs) as biomarkers in GC diagnosis [2].
Receptor tyrosine-protein kinase ErbB-3, is encoded by the ErbB3 gene and a member of the epidermal growth factor receptor (EGFR/ERBB) family of receptor tyrosine kinases [3]. The kinase-impaired ErbB3 is known to form active heterodimers with other members of the ErbB family, most notably the ligand binding-impaired ErbB2 [4]. ErbB3 was reported to be overexpressed in human gastric cancer, it acts as a tumor promoter by activation of a serial of complicated signaling including PI3K/AKT, RAS/RAF/MAPK and etc. The diversity of expression of ErbB3 was related to its upstream regulation, among all this mechanism miRNA regulation was one of the important reasons. miR-125, miR-199a, miR-205, and miR-450 etc. were all reported to potentially capable of regulating ErbB3, generally by targeting its three prime untranslated regions (3′UTR) [5,6,7,8,9].
MicroRNAs (miRNAs) are a class of small, noncoding, approximately 22-nucleotides-long RNAs, which may function as a post-transcriptional regulator of gene expression. Single nucleotide polymorphism (SNP) is a variation in a single nucleotide that occurs at a specific position in the genome, where each variation is present to some appreciable degree within a population. It commonly occurs in the human genome, some SNP were functional associating with the structure of functional proteins. However, SNP also occurred in 3′UTR of certain genes which might effect on the binding force by various miRNA [10, 11].
Thus, in this study, we focused on the SNPs in the 3′UTR of ERBB3. By using the bioinformatics software (http://bioinfo.life.hust.edu.cn/miRNASNP/), we obtained all the SNPs which could regulate by miRNAs (Table 2). Via the bioinformatics prediction and statistical analysis, we found that the rs3202538 (G/T) in ErbB3 3′-UTR might potential effect on the regulation by miR-204 and miR-211. Moreover, a previous study has revealed that rs3202538 (G/T) in ErbB3 3′-UTR was seriously related to type I diabetes just via regulation of miR-204 and miR-211 [12], we further investigated the allele distribution in a case–control study.
Materials and methods
Study subjects
A total of 851 GC cases and sex plus age-matched 799 controls obtained from Zhenjiang Hospital of Chinese Traditional Medicine, were included in this study. Patients were consecutively recruited between February 2010 and January 2015. All cases are incident ones during enrollment of the current case–control study. The diagnosis of all patients was histological confirmed. A face-to-face questionnaire was administered to collect demographic data and environmental exposure information, including alcohol use and cigarette consumption status as well as family cancer history. The normal tissues sampled was obtained at least 2 cm away from the margin of the tumor. All participants have provided their written informed consents to participate in this study. This study was approved by the Institutional Review Board of Zhenjiang Hospital of Chinese Traditional Medicine.
Genotype
The polymorphism was genotyped through the PCR-restriction fragment length polymorphism (RFLP) method as described previously [13]. The PCR reactions were carried out in a total volume of 5 μL containing TaqMan Universal Master Mix, 80X SNP Genotyping AssayMix, DNase-free water and 10 ng genomic DNA. The PCR conditions were 2 min at 50 °C, 10 min at 95 °C, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min by the 384-well ABI 7900HT Real Time PCR System. A 10% random sample was reciprocally examined by different persons, and the reproducibility was 100%.
Real-time PCR assay
Real-time polymerase chain reaction (RT-PCR) was performed to determine whether the mutation changed the expression level of ERBB3. The primers used for amplification were forward primer: CAGCAGCTTGACACACGGTA, and reverse primer: AAACACCAAAGTGGCATGTGA for ErbB3 and forward primer: TGTGGGCATCAATGGATTTGG, reverse primer: ACACCATGTATTCCGGGTCAAT for GAPDH. The amplification conditions were 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s, 55 °C for 40 s, and 72 °C for 30 s, and finally 4 °C for 30 min for cooling by the 384-well ABI 7900HT Real Time PCR System.
Cell lines and cell culture
GC cell lines SGC-7901 and MKN-45 were purchased from the Chinese Academy of Sciences Cell Bank. All cells were cultured in Dulbecco’s Minimum Essential Medium (DMEM) purchased from Gibco (CA, USA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, USA) and grown in humidified 5% CO2 at 37 °C. MiR-214 and miR-1225a mimics and a normal control were obtained from Genepharma (Shanghai, China). The transfection was conducted by using Lipofectamine 2000 (Invitrogen Corp, CA, USA).
Cell proliferation assays
Cell proliferation was determined by using CCK-8 (Dojin Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. Briefly, the control and infected cells were seeded at a density of 1 × 103 cells/well in 96-well plates. 10 μL of CCK-8 was added to each well containing 100 µL of the culture medium, and the plate was incubated for 2 h at 37 °C. The viability of cells was evaluated by measuring the absorbance at 450 nm, using a microplate reader (Thermo Labsystems, CA).
Western blot
For western blotting, proteins were extracted from tissues or cultured cells using RIPA buffer containing Protease Inhibitor Cocktails (P8340) (Sigma-Aldrich). An equal amount of proteins (100 μg) were separated with 7.5%/12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membrane. Primary polyclonal antibodies targeting ErbB3 (ab20161), p-AKT (ab38449), AKT (ab8933), β-actin (ab3280) were purchased from Abcam (Cambridge, MA). The secondary antibodies were anti-rabbit or anti-mouse HRP-linked were purchased from Santa Cruz Biotechnology (CA, USA). The blots were developed using ECL reagent (Millipore, MASS, USA). An equal amount of protein loading in each lane was confirmed using β-actin antibody. ImageJ software quantified the integrated density of the band.
Prediction of miRNAs binding to the SNP
Based on our bioinformatics analysis by using the bioinformatics software (http://www.bioguo.org/miRNASNP/) to predict the related SNPs in the 3′UTR of ERBB3 which could interact with miRNAs.
Construction of luciferase-based reporter plasmids
A full-length fragment of the 3′UTR containing rs3202538 (G/T) (wild type/mutant) were amplified. The PCR product was cloned into the pGL3-promoter luciferase-based plasmid (Promega) at the cloning site between KpnI and XhoI. The amplified fragment was verified by DNA sequencing. For cell proliferation and invasion assay, the full-length cDNA of ErbB3 was sub-cloned into the pGL3-ErbB3 3′UTR plasmid.
Immunohistochemistry
Sections were de-paraffinized and followed by rehydration steps through a graded ethanol series and distilled water, and then were treated with 3% H2O2 in methanol for 30 min to block the endogenous peroxidase activity. The sections were rinsed in phosphate-buffered saline (PBS) twice, 5 min each time and incubated with 10% normal goat serum for 30 min to block non-specific antibody binding. After washing, the samples were incubated with primary anti-rabbit antibody ErbB3 (ab16901) purchased from Abcam (Cambridge, MA) at 4 °C overnight, and then washed with PBS for three times and then incubated with secondary antibodies. After that, the sections were stained with DAB according to manufacturer’s protocols and mounted and photographed using a digitalized microscope camera (Nikon, Tokyo, Japan).
Dual-luciferase reporter assay
For luciferase activity analysis, SGC-7901 and MKN-45 cells were cotransfected with 100 ng of luciferase reporter constructs 5 ng of the β-gal control plasmid and 10 pmol of miRNAs with 1 µL Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen, NY, USA). After incubation for 48 h, we carried out the luciferase assay using the luciferase reporter assay system (Promega, Madison, WI) according to the manufacturer’s protocol. Measurements of luminescence and absorbance of β-gal were performed on a luminometer (Glomax 20/20; Promega). Three independent experiments were performed in triplicate.
Cell invasion assay
For trans-well assay, the chamber was treated with Matrigel before cells were inoculated, 100 μL cell suspension with serum-free medium was seeded to the upper chamber, cells were stained with crystal violet staining solution (Beyotime, Nantong, China). Migrated cells were counted by using Image-pro Plus 6.0 while cell numbers of the normal control group were normalized to 1.
Statistical analysis
Differences between cases and controls were evaluated by the Student’s t test for continuous variables and the χ2 test for categorical variables. The association between SNPs and GC risk was estimated by the OR and 95% CI using the general genetic model. The potential gene–environment interaction was evaluated by logistic regression analysis and tested by comparing changes in deviance between the models of main effects with or without the interaction term. The overall survival and relapse free survival rate in different groups were analyzed by using Kaplan–Meier curve. Comparisons between groups were analyzed by the t test (two-sided). All statistical analyses were performed using Prism Graphpad software.
Results
SNP rs3202538 in 3′UTR region of ErbB3 was a risk factor for development of gastric cancer
The differences in the distribution of the selected variables among GC and controls cases are listed in Table 1. No significant differences in age (P = 0.681) and sex (P = 0.286) were found between the case and the control group. There were more patients who had Hp infection in the GC patients than in the controls (P < 0.0001). However, all the variables above were further adjusted for any residual confounding effect in the later multivariate logistic regression analysis.
To investigate the miRNA associated SNPs in the 3′UTR of ERBB3, we first found all the possible SNPs from the SNP databases NCBI db SNP BUILED 129 and ENSEMBL v58 in the 3′-UTR of ErbB3 gene with the minor allele frequency (MAF) >0.05. And then used bioinformatics software Diana-Micro and RNA hybrid to predict miRNAs that can bind to the ErbB3 3′-UTR (data not shown). Finally, we obtained five SNPs in 3′UTR of ErbB3 which could be regulated by different miRNAs (Table 2). The positions of the SNPs in 3′UTR of ERBB3 as well as the variants were also listed. Further genotyping was performed to detect the distribution of allele gene of the SNP in our research, among all this SNPs, rs3202538 which can be potentially regulated by miR-204 miR-211 and miR-4278 were in Hardy–Weinberg equilibrium distribution pattern in the healthy control group (P < 0.0001).
Logistic regression analyses indicated that individuals with the GT and TT of rs3202538 in 3′UTR of ErbB3 were significantly associated with GC risk (P < 0.0001), which indicated that this SNPs might be a risk factor in GC development. Moreover, the significant association with GC risk was shown in U carrier of both SNPs (P < 0.0001) (Table 3). For SNP rs3202538, besides Wild type GG, other SNPs in 3′UTR of ErbB3 were significantly associated GC risk, especially for TT genotype (for TT: OR 4.32, 95% CI 1.34–1.88; for GT: OR 1.89, 95% CI 1.48–2.01; and for G carrier: OR 3.75; 95% CI 1.47–2.04). All ORs were adjusted for sex, age, and Hp. Infection status, or family cancer history.
SNP rs3202538 in 3′UTR region of ErbB3 was associated with GC clinical characters by deregulation of miR-204 and miR-211
Then, we did a stratified analysis of the association of the rs3202538 with the clinic pathological parameters of GC (Table 4). We found a significant association of the both genotypes with the tumor size, differentiation and metastasis as well as. Compared with the wild type, the carriers of T genotype presented significant large tumor size, poor differentiation as well as the high potential of metastasis (Table 4).
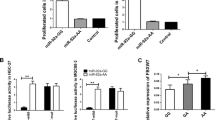
To investigate whether the rs3202538 SNPs affects the predicted miRNAs combining with ErbB3 3′UTR (Fig. 1a), we performed transient transfection in vitro and measured the relative activities with a Dual-Glo Luciferase Reporter Assay System. As it was indicated, co-transfection of the luciferase vector containing either wild type (GG) and homozygous T mutant 3′UTR of ErbB3 plus miR-204, miR-211 and miR4287 mimics and control into two GC cells, significantly increase the luciferase expression level was found in the mutant compared to wild types in both miR-204 and miR-211 but not miR-4087 (Fig. 1b). So it is indicated that the SNP in 3′UTR of ERBB3 can affect the binding affinity of either miR-204 and miR-211, and then affect the function of post-transcriptional regulation, resulting in the abnormal expression level of ErbB3.
SNP rs3202538 in ERBB3 3′UTR of GC patients with GT/TT genotype can up-regulate of ERBB3 transcription by deregulated by miR-211 and miR-204. a Bioinformatics predicted the binding site of the miR-204, miR-211 and miR-4287 with ErbB3 and the mutation types were conducted into the pGL3 plasmid as presented. b Cells were cotransfected with miR-204, miR-211 and miR-4287 mimics or control, Renilla luciferase vector pRL-SV40 for 48 h. Both firefly and Renilla luciferase activities were measured in the same sample. Firefly luciferase signals were normalized with Renilla luciferase signals. Data were presented as the mean ± SEM. Asterisk indicates a significant difference (P < 0.05)
G>T of rs3202538 promote cell proliferation and invasion by enhanced ErbB3 expression in miR-211 or miR-204 overexpression GC cell
To investigate the effect of G>T SNP on cell proliferation and metastasis, we overexpressed ErbB3 in MKN45 cells using the pGL3 vector, overexpression of ErbB3 can significantly increase the cell proliferation and cell invasion compared to the WT control. Both miR-211 and miR-204 can significantly decrease the cell proliferation and invasion ability in the G allele 3′UTR even lower than the WT control. However, there is no significant difference in both cell proliferation and invasion between the ErbB3 overexpressed MKN45 and MKN45 overexpressed T allele 3′UTR regulated ErbB3. This result indicated that the post-transcriptional regulation by miR-204 and miR-211 can be attenuated dramatically due to the G>T SNP in 3′UTR of ErbB3 (Fig. 1a–d). We also detected the expression of ErbB3 as well as its classical downstream AKT signaling by using western-blot, the protein expression of ErbB3 and the phosphorylated AKT in the residue of T308 can be significantly decreased in the G allele 3′UTR regulated ErbB3 overexpression MKN cells regulated by both miR-204 and miR-211. However, miR-204 and miR-211 have no apparent effects on the expression of ErbB3 or phosphorylated T308 AKT was investigated in the MKN45 cell overexpressed T allele 3′UTR regulated ErbB3 (Fig. 2e).
G>T of rs3202538 promote cell proliferation and invasion by enhanced ErbB3 expression in miR-211 or miR-204 overexpression GC cell. a, b The cell proliferation was determined for 5 days of the cells treated differently indicated in the figures. c, d The cell metastasis capability of variously treated MKN45 cells were determined by using a trans-well based cell invasion assay. e The expression of ErbB3 as well as activation of AKT signaling pathway in cells treated differently were determined by using western-blot. Data were presented as the mean ± SEM. *P < 0.05 and **P < 0.01
SNP rs3202538 in 3′UTR region of ErbB3 was contributed to upregulation of human GC ErbB3 and associated with poor prognosis of post-surgery GC patients
We also confirmed the expression of ERBB3 in clinical samples with different genotypes of rs3202538. ErbB3 expression was detected in human GC by IHC, there are strong, medium, weak and negative staining in varying sections, GG group ERBB3 staining components were significantly different to that in GT and TT group in ErbB3 expression (high 14.6%, medium 48.2% weak 26.8% and negative 10.4% for GG group; and high 27.6%, medium 38.5%, weak 25.7% and negative 8.2% for GT; high 36.1%, medium 33.6% weak 24.1% and negative 6.2% for GG group, P < 0.001) (Fig. 3a, b). Real-time PCR further confirmed such difference in ERBB3 transcription, however, there is no significant difference in miR-204 and miR-211 expression between three groups (Fig. 3c–e).
SNP rs3202538 in ERBB3 3′UTR on the expression of ERBB3 in clinical samples. a Representative figures for IHC staining of ERBB3 in GC tumor section. b Comparison of a component of IHC staining in both in TT, GT and GG genotype GC patents. c, d, e The expression level of ERBB3, miR-211 and miR-204 were determined by real-time PCR in different genotypes GC patents
Among of total 851 GC patients we have 344 patients with follow up data of survival and 36 GC patients have found reoccurrence. Therefore, survival analysis was performed in GG and U carrier group. Firstly, the overall 5-year survival rate in the TT/GT group was lowest as 15.72% which significantly different to GG group with a higher survival rate of 35.38% (P = 0.0004, HR = 2.045, 95% CI 1.321–2.644) (Fig. 4a, Additional file 1). The relapse free survival (RFS) was also investigated by Kaplan–Meier curve, and it is similar to the OS analysis that RFS in GG is also significantly higher than the T carrier group (P = 0.0004, HR = 2.059, 95% CI 1.329–2.659) (Fig. 4b, Additional file 2). In general, our foundlings indicated that the G/T SNP might serve as a tumor promoter and poor prognosis indicator in GC by affecting the binding of miR-204 and miR-211 on the 3′UTR of ErbB3.
Discussion
Overexpression of ErbB3 was reported in human gastric cancer in several independent types of research, and also a high expression of ErbB3 was regarded as an indicator of poor prognosis in post-surgery patients [3, 4, 14]. Recent works have implied the role of ErbB3 in gastric cancer as a key signaling hub [15]. On the one hand, HER3 overexpression may promote tumor progression and invasion by activation of PI3K/AKT signaling pathway [16]. On the other hand, a lot of direct evidence has emerged the benefit of anti-HER3 agents in combination with EGFR tyrosine kinase inhibitors as well as anti-HER2 agents in gastric cancer [17]. And moreover, alternative activation of c-MET mediated signaling was also dependent on ErbB3, which reflected as the development of resistance to c-MET inhibitors may result from the overexpression of ErbB3 [18].
The expression of ErbB3 was diverse in different GC patients, therefore, the signaling activation was also different. So far there was rarely research addressing the diversity of expression of ErbB3 in human GC, we addressed this issue here as SNP in its 3′UTR region and might cause deregulation of certain miRNA. miR-204 and miR-211 were found to have a potential suppression effect on ErbB3 transcription and SNP rs3202538 from G to T might increase ErbB3 expression by actuating such suppressive effect. In the present study, the difference of ErbB3 expression was verified between the G and T allele, the expression of ErbB3 can be attenuated dramatically by both miR-204 and miR-211 even in exogenous ErbB3 overexpression cells regulated by G allele but not the T allele 3′UTR. Also, the cell proliferation, invasion ability as well as downstream pathway activation also decreased corresponding to the expression of ErbB3.
The roles of miR-211 were complicated and controversial. Some of the researchers implied it as a tumor suppressor by targeting oncogenic genes such as IGF2R, TGFBR2 and NFAT5 in melanoma [19]. And moreover, in human breast cancer, miRNA-211 directly inhibit CDC25B expression in breast cancer cells, alters other related target proteins CCNB1 and FOXM1, and then inhibits breast cancer cells growth, migration, and invasion and lead G2/M arrest [20]. However, some studies defined miR-211 as a tumor promoter, by inhibiting the expression of SRC kinase signaling inhibitor 1 (SRCIN1), miR-211 was proved to promote human non-small cell lung cancer [21], and by targeting tumor suppressive gene CHD 5, miR-211 can promote human colon cancer [22, 23]. However, our study indicated that mIR-211 as a tumor suppressor in human GC by targeting ErbB3.
Compared to miR-211, the role of miR-204 was more explicit. In 2014 Zhou et al. reported that Hp infection can down-regulate expression of miR-204 and thus to promote human GC by targeting SOX4 [24], And miR-204 was extensively reported to suppression of EMT by targeting varying genes including snail and sirt1 [25, 26]. And in the present study, we revealed that miR-204 can also relate to metastasis of human GC by targeting ErbB3.
Conclusion
In the present study, we found miR-211 and miR-204 have a potential transcriptional suppressive effect on ErbB3, and SNP in 3′UTR region of ErbB3 can effectively associate significantly up-regulation of ErbB3 which might occur due to loss regulation of miRNA. These findings are novel and might contribute to explicit the individual differences in GC susceptibility.
Abbreviations
- GC:
-
gastric cancer
- SNP:
-
single-nucleotide polymorphism
- 3′UTR:
-
three prime untranslated region
- 95% CI:
-
95 confidence interval
- HR:
-
hazard ratio
- OD:
-
odds ratio
- CC:
-
CC genotype
- AA:
-
AA genotype
- C/A:
-
SNP C to A
References
Gastric Cancer Treatment (PDQ(R)). Health professional version. PDQ cancer information summaries. Bethesda (MD). 2002.
Li L, Wang H. Heterogeneity of liver cancer and personalized therapy. Cancer Lett. 2016;379:191–7.
Montemurro F, Scaltriti M. Biomarkers of drugs targeting HER-family signalling in cancer. J Pathol. 2014;232:219–29.
Roskoski R Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34–74.
Letelier P, Garcia P, Leal P, et al. miR-1 and miR-145 act as tumor suppressor microRNAs in gallbladder cancer. Int J Clin Exp Pathol. 2014;7:1849–67.
Momose K, Minami A, Shimono Y, et al. miR-214 and hypoxia down-regulate Necl-2/CADM1 and enhance ErbB2/ErbB3 signaling. Genes Cells. 2013;18:195–202.
Wang S, Huang J, Lyu H, et al. Functional cooperation of miR-125a, miR-125b, and miR-205 in entinostat-induced downregulation of erbB2/erbB3 and apoptosis in breast cancer cells. Cell Death Dis. 2013;4:e556.
Wu H, Mo YY. Targeting miR-205 in breast cancer. Expert Opin Ther Targets. 2009;13:1439–48.
Yu J, Li Q, Xu Q, et al. MiR-148a inhibits angiogenesis by targeting ERBB3. J Biomed Res. 2011;25:170–7.
Koutras A, Kotoula V, Fountzilas G. Prognostic and predictive role of vascular endothelial growth factor polymorphisms in breast cancer. Pharmacogenomics. 2015;16:79–94.
Mishra PJ, Mishra PJ, Banerjee D, et al. MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: introducing microRNA pharmacogenomics. Cell Cycle. 2008;7:853–8.
de Jong VM, Zaldumbide A, van der Slik AR, et al. Post-transcriptional control of candidate risk genes for type 1 diabetes by rare genetic variants. Genes Immun. 2013;14:58–61.
Sentinelli F, Filippi E, Fallarino M, et al. The 3′-UTR C>T polymorphism of the oxidized LDL-receptor 1 (OLR1) gene does not associate with coronary artery disease in Italian CAD patients or with the severity of coronary disease. Nutr Metab Cardiovasc Dis. 2006;16:345–52.
Amin DN, Sergina N, Lim L, et al. HER3 signalling is regulated through a multitude of redundant mechanisms in HER2-driven tumour cells. Biochem J. 2012;447:417–25.
Wang L, Yuan H, Li Y, et al. The role of HER3 in gastric cancer. Biomed Pharmacother. 2014;68:809–12.
Wu G, Qin XQ, Guo JJ, et al. AKT/ERK activation is associated with gastric cancer cell resistance to paclitaxel. Int J Clin Exp Pathol. 2014;7:1449–58.
Han S, Meng Y, Tong Q, et al. The ErbB2-targeting antibody trastuzumab and the small-molecule SRC inhibitor saracatinib synergistically inhibit ErbB2-overexpressing gastric cancer. MAbs. 2014;6:403–8.
Yun C, Gang L, Rongmin G, et al. Essential role of Her3 in two signaling transduction patterns: Her2/Her3 and MET/Her3 in proliferation of human gastric cancer. Mol Carcinog. 2015;54:1700–9.
Levy C, Khaled M, Iliopoulos D, et al. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol Cell. 2010;40:841–9.
Song GQ, Zhao Y. MicroRNA-211, a direct negative regulator of CDC25B expression, inhibits triple-negative breast cancer cells’ growth and migration. Tumour Biol. 2015;36:5001–9.
Ye L, Wang H, Liu B. miR-211 promotes non-small cell lung cancer proliferation by targeting SRCIN1. Tumour Biol. 2016;37:1151–7.
Cai C, Ashktorab H, Pang X, et al. MicroRNA-211 expression promotes colorectal cancer cell growth in vitro and in vivo by targeting tumor suppressor CHD5. PLoS ONE. 2012;7:e29750.
Naraparaju K, Kolla V, Zhuang T, et al. Role of microRNAs in epigenetic silencing of the CHD5 tumor suppressor gene in neuroblastomas. Oncotarget. 2016;7:15977–85.
Zhou X, Li L, Su J, et al. Decreased miR-204 in H. pylori-associated gastric cancer promotes cancer cell proliferation and invasion by targeting SOX4. PLoS ONE. 2014;9:e101457.
Liu Z, Long J, Du R, et al. miR-204 regulates the EMT by targeting snai1 to suppress the invasion and migration of gastric cancer. Tumour Biol. 2016;37:8327–35.
Zhang L, Wang X, Chen P. MiR-204 down regulates SIRT1 and reverts SIRT1-induced epithelial–mesenchymal transition, anoikis resistance and invasion in gastric cancer cells. BMC Cancer. 2013;13:290.
Authors’ contributions
Conception and design: SXC and SH. Collection and assembly of data: CX and YXW; Data analysis and interpretation: OYJ and HCX. Contribution of reagents, materials, and analysis tools, wrote the paper: QYC, OYJ and WDF. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
12935_2017_449_MOESM1_ESM.xlsx
Additional file 1. The output files of overal survival rate comparison in different groups analyzed by Kaplan–Meier survival curve.
12935_2017_449_MOESM2_ESM.xlsx
Additional file 2. The output files of relapse free survival rate comparison in different groups analyzed by Kaplan–Meier survival curve.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Shi, Y., Chen, X., Xi, B. et al. SNP rs3202538 in 3′UTR region of ErbB3 regulated by miR-204 and miR-211 promote gastric cancer development in Chinese population. Cancer Cell Int 17, 81 (2017). https://doi.org/10.1186/s12935-017-0449-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-017-0449-z