Abstract
Background
Anthocyanins are water-soluble flavonoids in plants, which give plants bright colors and are widely used as food coloring agents, nutrients, and cosmetic additives. There are several limitations for traditional techniques of collecting anthocyanins from plant tissues, including species, origin, season, and technology. The benefits of using engineering microbial production of natural products include ease of use, controllability, and high efficiency.
Results
In this study, ten genes encoding enzymes involved in the anthocyanin biosynthetic pathway were successfully cloned from anthocyanin-rich plant materials blueberry fruit and purple round eggplant rind. The Yeast Fab Assembly technology was utilized to construct the transcriptional units of these genes under different promoters. The transcriptional units of PAL and C4H, 4CL and CHS were fused and inserted into Chr. XVI and IV of yeast strain JDY52 respectively using homologous recombination to gain Strain A. The fragments containing the transcriptional units of CHI and F3H, F3’H and DFR were inserted into Chr. III and XVI to gain Strain B1. Strain B2 has the transcriptional units of ANS and 3GT in Chr. IV. Several anthocyanidins, including cyanidin, peonidin, pelargonidin, petunidin, and malvidin, were detected by LC-MS/MS following the predicted outcomes of the de novo biosynthesis of anthocyanins in S. cerevisiae using a multi-strain co-culture technique.
Conclusions
We propose a novel concept for advancing the heterologous de novo anthocyanin biosynthetic pathway, as well as fundamental information and a theoretical framework for the ensuing optimization of the microbial synthesis of anthocyanins.
Similar content being viewed by others
Introduction
Anthocyanins are water-soluble pigments in the form of glycosylated flavonoids, which produce the red to blue colors in fruits, vegetables, and grains [1]. Anthocyanins and anthocyanidins are frequently employed as food additives because of their vivid hues, nutritional values, and health function [2]. In addition, consumption of anthocyanin-rich foods could reduce the incidence of diseases in the circulatory system [3, 4], endocrine system [5, 6], digestive system [7], urinary system [8], nervous system [9], immune system [10], and ocular disorders [11], as well as play a role as an anticancer agent through the inhibition of inflammation and the reduction of oxidative stress [12]. The traditional method of obtaining anthocyanins is to extract them from plant tissues [13]. However, anthocyanins extracted from plants have the problems of seasonal and regional fluctuations in content, difficulty in controlling quality, and relatively low purity [14, 15]. Microbial synthesis of secondary metabolites has the advantages of being fast-growing, easy to culture, simpler, controllable, and cost-effective. Microbial cell factories are currently the most promising system for increasing anthocyanin production, using metabolic and genetic engineering methods to optimize genes in the synthetic pathway or directly transfer them into host cells to achieve anthocyanin production [16]. The use of microorganisms for anthocyanin biosynthesis is potentially advantageous.
To date, anthocyanins have been discovered in more than one thousand different natural varieties. The most prevalent and prolific anthocyanidins are six major classes: Pg (pelargonidin), Cy (cyanidin), Pn (peonidin), Dp (delphinidin), Pt (petunidin), and Mv (malvidin) [17]. Because of containing abundant anthocyanins, blueberries, one of the most commonly consumed berries with high nutritional value and several biological properties, have various health benefits for humans. The purple eggplant, one of the the Solanaceous vegetables, is more popular for the rich anthocyanins in the fruit skin. However, few studies have used the anthocyanins biosynthetic genes of blueberry and eggplant for heterologous synthesis of anthocyanins in microorganisms.
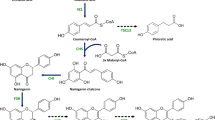
The anthocyanins biosynthetic genes of blueberry could be used for anthocyanins heterologous synthesis in microorganisms. The primary phases of anthocyanin manufacturing include the creation of flavonoid precursors, dihydro flavanone alcohols, and a variety of anthocyanins (Fig. 1). The most important synthetic enzymes in the role of creating the fundamental components of flavonoid compounds are phenolalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumaroyl-CoA ligase (4CL) [18]. Chalcone synthase (CHS) and chalcone isomerase (CHI) are involved in the synthesis of flavanones, which follow the phenylalanine synthesis pathway [19]. Flavonoid 3-hydroxylase (F3H), flavonoid 3’-hydroxylase (F3’H), and flavonoid 3’-5’-hydroxylase (F3’5’H) catalyze a series of hydroxylation reactions to generate dihydroflavanols [20], and these are then reduced to unstable intermediates through dihydroflavonol reductase (DFR) to produce leucoanthocyanidins [21]. which are converted to anthocyanidin in the form of glycosides by anthocyanidin synthase (ANS) and then glycosylated by 3-O-glycosyl transferase (3GT) [22]. Under certain conditions, these hydroxyl groups are further methylated by O-methyltransferase (OMT) to produce other anthocyanins such as peonidin 3-O-glucoside, petunidin 3-O-glucoside, and malvidin 3-O-glucoside (Fig. 1) [23].
Anthocyanin biosynthesis pathway in plants. Bold arrows indicate the synthetic routes in this study. Different colors represent different stages of anthocyanin synthesis. Yellow: synthesis stage of flavonoid precursors; pink: synthesis stage of dihydro flavanone alcohols; other colors: synthesis stages of various anthocyanins and anthocyanidins. The boxes on the right display the names of synthetic enzymes
The eukaryotic nature of Saccharomyces cerevisiae as a metabolic engineering platform for anthocyanin production may have the potential to promote the functional expression of plant-derived genes, in addition to its well-established synthetic biology technology. As a unicellular eukaryotic microorganism, S. cerevisiae has the advantages of simple cultivation conditions and rapid reproduction [24]. In contrast to the prokaryotic expression system, it can undergo post-transcriptional and translational modifications so that the expressed heterologous protein is structurally and functionally similar to the natural protein. One report showed that 400 µM naringenin was obtained from S. cerevisiae strain IMX106 in the culture medium [25]. Furthermore, by integrating fourteen additional genes into the yeast genome, and deleting three endogenous genes from the genome, the pelargonidin reached 0.01 µmol/gCDW in yeast intracellular with aerated, pH-controlled batch reactors [26]. By multiple strategies, a high-yield kaempferol-producing yeast strain was obtained with 86 mg/L of kaempferol. Meanwhile, 220 mg/L of naringenin and 200 mg/L of mixed flavonoids were achieved by two yeast strains’ co-culture system and supplementation of Tween 80 surfactants in the culture medium [27]. Another report developed a brewer’s yeast co-culture platform to enhance the production of naringenin (144.1 mg/L), kaempferol (168.1 mg/L), cornflower (31.7 mg/L), geraniol (33.3 mg/L), and delphinidin (15.8 mg/L) [28]. This co-culture strategy offers the possibility to efficiently and economically utilize microorganisms for anthocyanin production. Although several studies have achieved anthocyanin biosynthesis in S. cerevisiae, there are few reports about synthetic genes from blueberry used for anthocyanin heterologous biosynthesis in yeast.
In the study, the entire de novo anthocyanin synthetic pathway, including eight genes (C4H, 4CL, CHS, CHI, F3H, DFR, ANS, and 3GT) from blueberry (Vaccinium corymbosum), and two genes (PAL and F3’H) from purple round eggplant (Solanum melongena), were integrated into the S. cerevisiae different chromosomes by homologous recombination (HR) to construct three yeast strains (Strain A, B1 and B2). The production of different anthocyanins and anthocyanidins was successfully realized in yeast using glucose as the substrate by coculture and pH control.
Materials and methods
Genes, strains, plasmids, and media
All genes, strains, and plasmids used in this study are listed in Additional file 1: Table S1 and the primers used here are listed in Additional file 1: Table S2 and Table S3. Highbush blueberry and purple round eggplant were used for five anthocyanin synthetic genes cloning. Genome integration was performed by HR in S. cerevisiae JDY52 using the POT vector. Yeast host strain JDY52-URR1-His-URR2 (MATahis3Δ200 leu2Δ0lys2Δ0trp1Δ63 ura3Δ0 met15Δ0) was donated by Dr. Junbiao Dai of Tsinghua University [29].
All plasmid-containing E. coil DH5α strains were cultured in Luria - Bertani (LB) broth with shaking at 37°C. Yeast was cultured in YPD medium with shaking at 30°C. Recombinant yeast was cultured in SD nutrient-deficient medium (containing 20 g/L glucose, 6.7 g/L YNB), replenished with 10× amino acid triple-deficient master batch and 100×LEU/URA/TRP masterbatch.
Cloning of the anthocyanin synthetic genes
The mRNA was extracted from blueberry and eggplant using the Trizol Method. The first strand cDNA was synthesized by Reverse Transcription Kit (Yisheng, Shanghai) and stored at -20℃. The PAL, C4H, 4CL, CHS, and F3’H were amplified (Additional file 1: Table S2), ligated with a T-vector (Tiangen, Beijing), and transformed into E. coli DH5α. The positive bacteria verified by PCR was expanded and cultured, and then the plasmid was extracted, identified, and sequenced.
Construction of transcription units of anthocyanin synthetic genes
The anthocyanin synthetic genes, PAL, C4H, 4CL, CHS, and F3’H obtained by cloning, CHI, F3H, DFR, ANS, and 3GT stored at the lab [30], were used to construct complete transcription units, including promoters, CDS and terminator, by the Yeast Fab Assembly method as previously described [31]. The vectors of HCKan-Pro (the promoter pf GPD, TEF2, ADH1, or ACS2), HCKan-T (the terminator of ADH1), and POT-RFP (used for carrying the transcriptional unit) were digested by Bsm BI (NEB, Beijing), and the open reading frames (anthocyanin synthetic genes) were digested by BsaI (NEB, Bejing). Then, due to the generation of specific sticky ends after enzymatic cleavage, transcription units (POT-TUs) are assembled to ligate promoters, open reading frames, terminators, and POT vectors in turn by T4 ligase (NEB, Beijing) (Fig. 2a). Finally, 6×His or HA tag (Additional file 1: Table S3) was added to the C-terminus of transcription units by overlap extension PCR for subsequent protein validation.
Construction of the tandem recombinant S. Cerevisiae strain
To construct recombinant yeast strains expressing entitled anthocyanin biosynthetic pathway independently of the maintenance medium, PAL and C4H, 4CL and CHS, CHI and F3H, F3’H and DFR, ANS and 3GT were linked in tandem and integrated into the chromosomes III, IV, and XVI of yeast respectively (Fig. 2b). Firstly, PMV- ADP1(PGK1)/URRs/SURTUTDs (the homologous arms) and PMV- LEU/URA/TRP (the selection marker) were digested with BsmBI (NEB), and different transcription units were digested with BsaI (NEB). Then, the five fragments, including the homologous arm (Left), transcription unit 1, transcription unit 2, selection marker, and homologous arm (Right), were sequentially ligated to one fragment by T4 DNA ligase (NEB) due to the specificity of cohesive ends, which was transformed to yeast strain JDY52 using the classical lithium acetate (LiAc) method [32] and recombinant yeast strains were enabled to grow on SD-LEU/URA/TRP dystrophic medium. Single colonies of recombinant S. cerevisiae strains purified by streaking on SD screening plates were picked in 3 mL of YPD and incubated in an incubator at 30°C and 180 rpm for 24 h. Yeast cells were centrifuged and genomic DNA [33] was extracted for detection by PCR.
Workflow diagram for constructing recombinant yeast strains. (a) Construction of the transcription units. Anthocyanin synthetic genes were digested by BsaI (NEB). The promoters (HCKan-PGPD/TEF2/ADH1/ACS2), terminator (HCKan-TADH1), and vector carrying the transcriptional unit (POT-RFP) were digested by BsmBI (NEB). Then, four fragments were ligated by T4 ligase to form the transcription unit. TUs: transcription units. (b) Construction of recombinant yeast strains. pMV-ADP1(PGK1)/URRs/SURTUTDs and pMV-LEU/URA/TRP were digested with BsmBI (NEB), and different transcription units were digested with BsaI (NEB). These five digested fragments were sequentially ligated to one fragment by T4 DNA ligase (NEB), which was transformed to yeast strain JDY52 using the classical LiAc method. HAs: homology arms, ADP1(PGK1)/URRs/SURTUTDs. AAs: amino acids, LEU/URA/TRP. Pn: promoter, n = 1, 2
Western blotting
The strains with correct genotype verification were delineated and purified for subsequent protein expression assays. Single colonies were picked in 3 mL of YPD, cultured overnight at 180 rpm 30℃ for 12 h, and then transferred to a triangular flask of 10 mL YPD at a ratio of 1: 50, and continued to be cultured at 180 rpm 30℃ for 48 h. The proteins in the supernatant were lysed, denatured at high temperature, and then subjected to electrophoresis on sodium dodecyl sulfate-polyacrylamide gels. After electrophoresis, the proteins were transferred onto a polyvinylidene fluoride membrane using rabbit anti-His (anti-HA) monoclonal antibody (Yeasen, Shanghai) diluted 1: 5000 as primary antibody and enzyme-labeled goat anti-rabbit IgG (Yeasen, Shanghai) antibody diluted 1: 5000 as secondary antibody. Signals were excited using Super Signal West Pico PLUS Chemiluminescent substrate (Bio-Rad, USA) and images were acquired using a Gel DocTM XR + imaging system (Bio-Rad, USA).
Quantitative analysis of anthocyanins
The recombinant S. cerevisiae strains (A, B1 and B2), obtained through the specified genetic manipulation, were inoculated into 3 mL of YPD medium in the ratio of 4: 2: 1 and incubated at 30 °C with shaking at 180 rpm for 24 h. As a crucial control, the original strain S. cerevisiae JDY52 was similarly inoculated and incubated under identical conditions (30 °C, 180 rpm for 24 h) in parallel. Following this initial incubation, yeast cells from both the recombinant strains and the control strain were collected by centrifugation and resuspended in 10 mL of yeast synthesis medium (containing 20 g/L glucose, 6.7 g/L YNB, 10 g/L (NH4)2SO4, 3 g/L KH2PO4, 0.24 g/L MgSO4, adjusted to pH 5.0). Subsequently, Dimethyl 2-oxoglutarate (0.1 mM) and sodium ascorbate (2.5 mM) were added to the suspensions. All cultures were further incubated for 48 h at 30 °C with shaking at 140 rpm. After this extended incubation period, the cultures were concentrated, with the performance of the recombinant strains being compared to that of the original JDY52 strain, which served as the primary control for assessing the effects of genetic modifications.
The quantitative analysis was carried out using an AB SCIEX QTRAP 6500PLUS MS/MS (AB Sciex, Framingham, MA, USA). Chromatographic separation of the analytes and IS was performed on an ACQUITY BEH C18 (1.7 μm, 2.1 × 100 mm) at 40℃. The temperature of the autosampler was maintained at 4 °C and 2 µL was injected into the system. The mobile phase of water (containing 0.1% formic acid, A) and acetonitrile (B) was performed at gradient elution with a flow rate of 0.2 mL/min: 0 min, 80% A, 20% B; 1 min, 80% A, 20% B; 3 min, 25% A, 75% B; 4 min, 25% A, 75% B; 4.05 min, 80% A, 20% B; 6.5 min, 80% A, 20% B; 7 min, 80% A, 20% B. The electrospray ion source temperature is 550 °C; the mass spectrometry voltage in positive ion mode is 4500 V; and the gas curtain gas pressure is 35 psi. In the LC-MS/MS coupler, each ion pair is scanned and detected based on optimized de-clustering voltages and collision energies.
Results
The genes cloning of anthocyanin biosynthetic pathway
Despite the abundance of anthocyanins in blueberries and their potential health benefits, cloning all the necessary genes for a complete anthocyanin biosynthetic pathway from blueberries alone proved to be a significant bottleneck. While we had successfully cloned five essential anthocyanin synthetic genes (CHI, F3H, DFR, ANS, and 3GT, stored in our laboratory) from blueberries, we encountered difficulties in isolating two crucial genes: phenylalanine ammonia-lyase (PAL) and flavonoid 3’-hydroxylase (F3’H). PAL is a key enzyme in the initial steps of the phenylpropanoid pathway, which feeds into the anthocyanin biosynthetic pathway. Similarly, F3’H plays a pivotal role in determining the color and pattern of anthocyanin pigmentation by catalyzing the hydroxylation of flavonoids at the 3’ position. Given the urgent need to complete the pathway, we turned to purple round eggplant as an alternative source. Eggplants, like blueberries, are rich in anthocyanins and possess a similar biosynthetic machinery. Through diligent efforts, we were able to clone PAL and F3’H from purple round eggplant, overcoming the cloning difficulties encountered with blueberries (Fig. 3). The incorporation of these two additional genes, along with the previously cloned five from blueberries and three others (C4H, 4CL, and CHS) also sourced from blueberry, allowed us to assemble a comprehensive set of ten genes capable of synthesizing two major types of anthocyanins: cyanidin and pelargonidin (Table 1).
Cloning of anthocyanin synthase genes PAL, C4H, 4CL, CHS, and F3’H. Three synthase genes C4H, 4CL, and CHS were amplified using blueberry cDNA as the template. Vc: blueberry (V. corymbosum). The cDNA of purple round eggplant was used as a template to amplify the remaining two synthase genes PAL and F3`H. Sm: purple round eggplant (S. melongena). M: DNA marker
The construction of transcription units
Using the Yeast Fab Assembly method, we successfully created transcription units for ten anthocyanin synthase genes. These units replaced the RFP region of the POT-RFP plasmid, enabling easy screening based on colony color: ligation failures appeared red, while successful colonies were white. Through colony PCR with gene-specific primers (Table S3), we validated and selected the white colonies, resulting in ten plasmids carrying the respective anthocyanin synthase genes: POT-PACS2-4CL, POT-PGPD-CHS, POT-PTEF2-PAL, POT-PADH1-C4H, POT-PADH1-CHI, POT-PTEF2-F3H, POT-P TEF2-F3’H, POT-PGPD-DFR, POT-PGPD-ANS, and POT-PACS2-3GT, were gained (Fig. 4a). The red/white spot screening technique significantly enhanced the accuracy of our selection process.
To facilitate protein expression analysis, we appended either a 6×His or HA tag to the C-terminal of each gene via PCR amplification. The construction details, protein sizes, and identified tags were shown as Table 2. After selecting a single clone for cultivation and plasmid extraction, PCR confirmed the presence of the correct tag (Fig. 4b), and sequencing verified the final plasmid constructs.
Transcription units construction and validation. (a) Plasmids construction diagram. The standard parts (Promoter, Ori, Terminator) and the “POT” acceptor vector are mixed with buffer and enzymes to assemble a transcription unit. (b) Verification results of adding 6×His or HA tag to the transcription unit of plasmids. The transcription units of PAL, C4H, 4CL, CHI, F3H, F3’H, DFR, 3GT, and ANS were constructed with a 6×His tag added to the C-terminus of these genes, and CHS was constructed with an HA tag added to the C-terminus of the target gene by Overlap PCR. The plasmid PCR validation of the genes together with tag using gene-specific primers. M: DNA marker
Construction of recombinant yeast strains
Using Yeast Fab Assembly, we successfully constructed complete transcriptional units for anthocyanin biosynthetic genes in vitro. The PAL and C4H transcriptional units were fused and integrated into Chr. XVI of S. cerevisiae JDY52 via HR, confirming the successful creation of recombinant yeast strain JDY52-PAL-C4H. PCR analysis verified the presence of the PAL target band (Fig. 5b), and Western Blot analysis confirmed the expression of PAL and C4H proteins in the strain (Fig. 5c). Furthermore, the transcriptional units of 4CL and CHS were fused and inserted into chromosome IV of JDY52-PAL-C4H, generating strain JDY52-PAL-C4H-4CL-CHS (Strain A). PCR confirmed the presence of the 4CL target band (Fig. 5d), and Western Blot analysis validated the expression of 4CL and CHS proteins in the strain (Fig. 5e). These results demonstrate the successful construction of Strain A, harboring multiple anthocyanin biosynthetic genes.
Construction and detection of yeast strain JDY52-PAL-C4H-4CL-CHS (Strain A). (a) Schematic diagram of constructing JDY52-PAL-C4H-4CL-CHS (Strain A). (b) PCR verified the PAL from the recombinant yeast JDY52-PAL-C4H. (c) The protein levels of PAL and C4H were detected in the strain JDY52-PAL-C4H by western blotting. (d) PCR verified the 4CL from the recombinant yeast JDY52-PAL-C4H-4CL-CHS. (e) The protein levels of PAL, C4H, and 4CL were detected in the strain JDY52-PAL-C4H-4CL-CHS by western blotting. (f) The protein levels of CHS were detected in the strain JDY52-PAL-C4H-4CL-CHS by western blotting
The results demonstrated successful construction of the recombinant yeast strain S. cerevisiae JDY52-CHI-F3H-F3’H-DFR (Strain B1), evidenced by PCR confirmation of F3H and F3’H integration and Western Blot verification of CHI, F3H, F3’H, and DFR protein expression (Fig. 6b-d). However, when integrating additional transcriptional units of ANS and 3GT into S. cerevisiae JDY52 to create Strain B2 (JDY52-ANS-3GT), PCR confirmed their insertion (Fig. 6e-f), but targeted protein bands were not detected by Western Blot, suggesting limited expression of these proteins. Notably, culturing these strains revealed that growth duration correlated with gene integration complexity, potentially indicative of expression inhibition due to high gene load. Despite attempts, the medium did not turn red, indicating inadequate anthocyanin production, likely due to low ANS and 3GT expression levels as seen in protein assays. This suggested the possibility of proanthocyanidins or other precursors being produced instead of anthocyanins in the yeast strains.
Construction and detection of yeast strain JDY52-CHI-F3H-F3’H-DFR (Strain B1) and JDY52-ANS-3GT (Strain B2). (a) Schematic diagram of constructing JDY52-CHI-F3H-F3’H-DFR (Strain B1) and JDY52-ANS-3GT (Strain B2). (b) PCR verified the F3H from the recombinant yeast JDY52-CHI-F3H. (c) PCR verified the F3’H from the recombinant yeast JDY52-CHI-F3H-F3’H-DFR. (d) The protein levels of CHI, F3H, F3’H, and DFR were detected in the strain JDY52-CHI-F3H-F3’H-DFR by western blotting. (e) PCR verified the ANS from the recombinant yeast JDY52-ANS-3GT. (f) PCR verified the 3GT from the recombinant yeast JDY52-ANS-3GT
Anthocyanins detection and analysis of fermentation broth by LC-MS/MS
To alleviate the problem of metabolic stress in single-strain fermentation and accurately assess the benefits of multi-strain co-culture for anthocyanin synthesis, this study employed a comprehensive control approach. Firstly, Strains A, B1, and B2 were individually pre-cultivated in YPD medium under standard conditions, serving as baselines for their individual growth and metabolic activities. Additionally, as a crucial control, the original strain S. cerevisiae JDY52 was also pre-cultivated in YPD medium for 24 h, mimicking the initial growth phase of the experimental strains. Subsequently, Strains A, B1, and B2 were inoculated into a single flask in the ratio of 4: 2: 1 for co-culture in YPD medium, supplemented with dimethyl 2-ketoglutarate and sodium ascorbate to enhance anthocyanin production. This co-culture was then concentrated and subjected to LC-MS/MS detection for quantitative analysis of anthocyanin content.Parallel to this, the original yeast JDY52 strain was also inoculated into a separate flask, fermented under identical conditions (YPD medium with dimethyl 2-ketoglutarate and sodium ascorbate) for 24 h, and processed in the same manner as the co-culture. The supernatant of fermentation broth was concentrated and analyzed by LC-MS/MS.
The anthocyanins present were identified in each peak by matching the m/z values, fragmentation patterns, and retention times. A total of twenty-seven peaks containing previously identified anthocyanins, many of which were differently modified, were found in fermentations by LC-MS/MS analysis (Additional file 1: Table S4). All of the six most common anthocyanidins, including cyanidin, peonidin, pelargonidin, delphinidin, petunidin, and malvidin were detected here (Additional file 1: Table S4). Meanwhile, based on the peak area results, cyanidin accounted for about 9.3% of the total anthocyanins in control group, which was elevated to 12.1% in co-culture strains. The peonidin, pelargonidin, petunidin, and malvidin were from 2.0%, 10.0%, 4.0%, and 19.5–6.8%, 28.1%, 21.2%, and 35.6% respectively. However, delphinidin was reduced from 54.7 to 11.1% (Additional file 1: Table S4). These data identified the anthocyanin biosynthetic pathway constructed in the yeast strain could increase the levels of anthocyanin by coculture. Furthermore, the delphinidin content was reduced may because F3’5’H was not cloned and integrated into the yeast genome in this study.
Furthermore, elevated relative intensities of six anthocyanidins, oenin chloride, callistephin chloride, pelargonidin O-acetylhexoside, peonidin chloride, cyanidin O-acetylhexoside, and pelargonidin O-acetylhexoside, were detected in the fermentation broth of recombinant yeast strains as compared to the original JDY52 strain (Fig. 7). Compared to JDY52 control cultures, the relative intensities of eight anthocyanins and anthocyanidins reduced in recombinant co-cultures, in recombinant strain cultures reduced, including malvidin 3,5-diglucoside, cyanidin 3-O-glucoside, delphinidin O-malonyl-malonylhexoside, petunidin 3-O-rutinoside, delphinidin 3-sophoroside-5-rhamnoside, delphinidin O-malonylhexoside, cyanidin O-malonyl-malonylhexoside, and pelargonidin 3-O-malonylhexoside (Fig. 8). There is enough evidence for the final synthesis of cyanidin, peonidin, pelargonidin, petunidin, and malvidin was catalyzed in the coculture of yeast recombinant strains by ten anthocyanin synthases, PAL, C4H, 4CL, CHS, CHI, F3H /F3’H, DFR, ANS, and 3GT. The successful synthesis of anthocyanins was identified by mass spectrometry, a more sensitive method, demonstrating the feasibility of synthesizing anthocyanins from scratch in S. cerevisiae.
LC-MS/MS analysis of the control JDY52 and recombinant yeast co-culture fermentation broth with elevated anthocyanin content. These types of anthocyanins and anthocyanidins increased in co-culture fermentation broth. The peak area of Oenin chloride (a), Callistephin chloride (c), Pelargonidin O-acetylhexoside (e), Peonidin chloride (g), Cyanidin O-acetylhexoside (i), and Pelargonidin O-acetylhexoside (k) produced in the fermentation broth of JDY52 (control). The peak area of Oenin chloride (b), Callistephin chloride (d), Pelargonidin O-acetylhexoside (f), Peonidin chloride (h), Cyanidin O-acetylhexoside (j), and Pelargonidin O-acetylhexoside (l) produced from the recombinant co-culture strains
LC-MS/MS analysis of the control JDY52 and recombinant yeast co-culture fermentation broth with reduced anthocyanin content. These types of anthocyanins and anthocyanidins were lower in co-culture fermentation broth than that of JDY52 strain. The peak area of Malvidin 3,5-diglucoside (a), Cyanidin 3-O-glucoside (c), Delphinidin O-malonyl-malonylhexoside (e), Petunidin 3-O-rutinoside (g), Delphinidin 3-sophoroside-5-rhamnoside (i), Delphinidin O-malonylhexoside (k), Cyanidin O-malonyl-malonylhexoside (m), and Pelargonidin 3-O-malonylhexoside (o) produced in the fermentation broth of JDY52 (control). The peak area of Malvidin 3,5-diglucoside (b), Cyanidin 3-O-glucoside (d), Delphinidin O-malonyl-malonylhexoside (f), Petunidin 3-O-rutinoside (h), Delphinidin 3-sophoroside-5-rhamnoside (j), Delphinidin O-malonylhexoside (l), Cyanidin O-malonyl-malonylhexoside (n), and Pelargonidin 3-O-malonylhexoside (p) produced from the recombinant co-culture strains
Discussion
Traditional methods for extracting anthocyanins from plant tissues are limited by plant growth, arable land constraints, and operationally difficult extraction methods, making it very expensive to obtain anthocyanins. Although metabolic engineering with microbes is a very promising approach, there are still challenges associated with the large-scale production of anthocyanins [34]. In this study, the de novo anthocyanin synthesis pathway was constructed in yeast using anthocyanin synthase genes from anthocyanin-rich blueberry and eggplant. We selected different promoters to regulate the expression of ten anthocyanin synthesis genes in constructing transcription units in vitro (Fig. 2a), and HR was carried out through different homology arms ADP1(PGK1), URRs, and SURTUTDs. Using the Yeast Fab Assembly method, the assembled complete transcription units were integrated into the target Chr. III, IV, and XVI, respectively. There were enzyme cleavage sites and unique nucleotide sequences of BsmBI added upstream and downstream of the homology arms and screening markers. The unique four-base sticky end will appear after the enzymatic cleavage, ensuring that the individual elements could be connected in passing, realizing the two-by-two tandem linkage of anthocyanin synthesis-related enzyme genes (Fig. 2b). In this study, the type IIs restriction endonuclease was used to realize the ligation of four or five fragments, and several fragments were digested and ligated at the same time under certain conditions, instead of being digested and then ligated by T4 ligase [35]. In advance, we realized the sequential joining of several fragments by designing unique sticky ends. In addition, the recombinant yeast vectors were constructed by different screening markers for subsequent cultures.
Although some anthocyanin synthase genes from several different plants have been used for constructing the anthocyanin synthase pathway in microorganisms, the genes of anthocyanin-rich blueberry and eggplant were not reported for the anthocyanin synthase through microbial fermentation to our knowledge. This study was initially envisioned to use anthocyanin-rich blueberry fruits exclusively as the experimental material, and it was proposed to amplify all genes of three main anthocyanin synthase pathways from blueberries, aiming to synthesize all six common anthocyanidins (Fig. 1). By maintaining the species uniformity of synthase genes to maintain the consistency of anthocyanin synthase activity. At the same time, blueberry fruits are rich in anthocyanins, which predicts a high activity of anthocyanin synthase (at certain times) in blueberries. However, in the actual research process, when blueberry cDNA was used as the template to amplify the PAL and F3’H, there was no purposeful band, and attempts to replace a variety of conditions failed to obtain the desired results. Finally, the pericarp cDNA of purple round eggplant was selected as the template to amplify the PAL and F3’H. Several attempts to amplify the F3’5’H resulted in no target bands, so the pathway strain where the F3’5’H is located was not constructed. Theoretically, Yeast Fab Assembly technology can realize the tandem of six transcriptional units [29]. However, in this study, the construction of the three-gene tandem unit of ANS, 3GT and OMT was unsuccessful in this study. So it was replaced by the construction of the tandem of ANS and 3GT, and successfully transformed yeast. When culturing recombinant yeast strains, it was found that the time required for yeast growth was prolonged as more genes were integrated into the genome. It was suspected that a single strain inhibited itself growth due to the integration of too many genes. So an attempt was made to integrate the tandem ANS and 3GT fragment alone into the JDY52 strain, and the JDY52-ANS-3GT (Strain B2) was constructed.
In yeast strains, the purposeful bands of ANS and 3GT were not detected. ANS catalyzed the conversion of colorless intermediates to colored anthocyanidins. However, at this point, the anthocyanidins produced were unstable [36]. 3GT subsequently catalyzes their conversion to more stable, colored anthocyanins and anthocyanidins [22]. Codon optimization of these two genes has been proposed in multiple studies to increase anthocyanins content [26]. In this study, codon optimization of the genes was not performed, which may have contributed to the non-expression of ANS and 3GT, or it may have been impossible to detect protein expression due to low expression levels or spatiotemporal specificity.
Although there were no purposeful bands for ANS and 3GT in the Western Blot assay, this did not rule out the possibility that they might pass the WB assay due to low expression, so an attempt at anthocyanin synthesis was made in this study. Ideally, the medium that produces anthocyanins and anthocyanidins would turn red, however, the medium color did not change during the experiment. Based on the results of the protein expression assay in recombinant yeast, the ANS and 3GT may not have been expressed, and it is suspected that anthocyanins were not produced. But proanthocyanidins or other colorless intermediates may have been produced, or there may also be cases where the amount of anthocyanins or anthocyanidins produced was too low to be recognized by changes in the color of the medium. Therefore, the yeast fermentation broth was analyzed by LC-MS/MS to determine if anthocyanin analogs were synthesized, and the results showed that five anthocyanidins increased in the recombinant yeast, providing sustainable implications for the production of anthocyanins in microorganisms.
Conclusion
We cloned ten anthocyanin synthase genes from blueberry or eggplant, and created complete transcriptional units for each of them, which were inserted into the different chromosomes of yeast to construct recombinant strains (A, B1 and B2). Ultimately, using the LC-MS/MS method, the increases of cyanidin, peonidin, pelargonidin, petunidin, and malvidin were identified in the broth extract of recombinant yeast fermentation compared with that of JYD52. The construction of heterologous de novo biosynthesis pathway for anthocyanins in S. cerevisiae will facilitate the production of anthocyanins through microbial fermentation. Our research offers fresh concepts for developing a de novo heterologous pathway for the biosynthesis of anthocyanins. It also offers fundamental information and a theoretical framework for optimizing the microbial synthesis of anthocyanins in the future.
Data availability
All data generated or analyzed during this study are included in this published article [and its Additional files].
References
Khoo HE, Azlan A, Tang ST, Lim SM. Anthocyanidins and anthocyanins: colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr Res. 2017;61:1361779. https://doi.org/10.1080/16546628.2017.1361779.
Zhang, Butelli. Martin. Engineering anthocyanin biosynthesis in plants. CURR OPIN PLANT BIOL. 2014;19:81–90. https://doi.org/10.1016/j.pbi.2014.05.011.
García-Conesa MT, Chambers K, Combet E, Pinto P, Garcia-Aloy M, Andrés-Lacueva C, de Pascual-Teresa S, Mena P, Konic Ristic A, Hollands WJ, et al. Meta-analysis of the effects of foods and Derived products containing ellagitannins and anthocyanins on cardiometabolic biomarkers: analysis of factors influencing variability of the individual responses. Int J Mol Sci. 2018;19:694. https://doi.org/10.3390/ijms19030694.
Igwe EO, Charlton KE, Probst YC. Usual dietary anthocyanin intake, sources and their association with blood pressure in a representative sample of Australian adults. J Hum Nutr Dietetics. 2019;32:578–90. https://doi.org/10.1111/jhn.12647.
Nemes A, Homoki JR, Kiss R, Hegedűs C, Kovács D, Peitl B, Gál F, Stündl L, Szilvássy Z, Remenyik J. Effect of Anthocyanin-Rich Tart Cherry Extract on Inflammatory mediators and adipokines involved in type 2 diabetes in a high Fat Diet InducedObesity Mouse Model. Nutrients. 2019;11:1966. https://doi.org/10.3390/nu11091966.
Qian X, Wang X, Luo J, Liu Y, Pang J, Zhang H, Xu Z, Xie J, Jiang X, Ling W. Hypouricemic and nephroprotective roles of anthocyanins in hyperuricemic mice. Food Funct. 2019;10:867–78. https://doi.org/10.1039/c8fo02124d.
Wang Y, Zhao L, Wang D, Huo Y, Ji B. Anthocyanin-rich extracts from blackberry, wild blueberry, strawberry, and chokeberry: antioxidant activity and inhibitory effect on oleic acid-induced hepatic steatosis in vitro. J Sci Food Agric. 2016;96:2494–503. https://doi.org/10.1002/jsfa.7370.
Li L, Li J, Xu H, Zhu F, Li Z, Lu H, Zhang J, Yang Z, Liu Y. The Protective Effect of anthocyanins extracted from Aronia Melanocarpa Berry in Renal Ischemia-Reperfusion Injury in mice. Mediat Inflamm. 2021;2021:7372893. https://doi.org/10.1155/2021/7372893.
Tikhonova MA, Shoeva OY, Tenditnik MV, Ovsyukova MV, Akopyan AA, Dubrovina NI, Amstislavskaya TG, Khlestkina EK. Evaluating the Effects of Grain of Isogenic Wheat Lines Differing in the content of anthocyanins in Mouse models of neurodegenerative disorders. Nutrients. 2020;12:3877. https://doi.org/10.3390/nu12123877.
Hyun KH, Gil KC, Kim SG, Park SY, Hwang KW. Delphinidin Chloride and its hydrolytic metabolite gallic acid promote differentiation of Regulatory T cells and Havean anti-inflammatory effect on the allograft model. J Food Sci. 2019;84:920–30. https://doi.org/10.1111/1750-3841.14490.
Nabavi SF, Habtemariam S, Daglia M, Shafighi N, Barber AJ, Nabavi SM. Anthocyanins as a potential therapy for diabetic retinopathy. Curr Med Chem. 2015;22:51–8. https://doi.org/10.2174/0929867321666140815123852.
Kalemba-Drozdz M, Cierniak A, Cichon I. Berry fruit juices protect lymphocytes against DNA damage and ROS formation induced with heterocyclic aromatic amine PhIP. J Berry Res. 2020;10:95–113. https://doi.org/10.3233/jbr-190429.
Arruda HS, Silva EK, Peixoto Araujo NM, Pereira GA, Pastore GM, Marostica Junior MR. Anthocyanins recovered from Agri-Food By-Products using innovative processes: trends, challenges, and perspectives for their application in Food systems. Molecules. 2021;26:2632. https://doi.org/10.3390/molecules26092632.
Zhu L, Huang Y, Zhang Y, Xu C, Lu J, Wang Y. The growing season impacts the accumulation and composition of flavonoids in grape skins in two-crop-a-year viticulture. J Food Sci Technology-Mysore. 2017;54:2861–70. https://doi.org/10.1007/s13197-017-2724-3.
Ryan JM, Revilla E. Anthocyanin composition of Cabernet Sauvignon and Tempranillo grapes at different stages of ripening. J Agric Food Chem. 2003;51:3372–8. https://doi.org/10.1021/jf020849u.
Marchev AS, Yordanova ZP, Georgiev MI. Green (cell) factories for advanced production of plant secondary metabolites. Crit Rev Biotechnol. 2020;40:443–58. https://doi.org/10.1080/07388551.2020.1731414.
Zha J, Wu X, Koffas MA. Making brilliant colors by microorganisms. Curr Opin Biotechnol. 2020;61:135–41. https://doi.org/10.1016/j.copbio.2019.12.020.
li LI, Yue Z, Junlan MA. Recent progress on key enzymes: PAL、C4H、4CL of phenylalanine metabolism pathway. China J Bioinf. 2007;5:187–9.
Sun W, Shen H, Xu H, Tang X, Tang M, Ju Z, Yi Y. Chalcone Isomerase a key enzyme for anthocyanin biosynthesis in Ophiorrhiza Japonica. Front Plant Sci. 2019;10:865. https://doi.org/10.3389/fpls.2019.00865.
Xu S, Li G, Zhou J, Chen G, Shao J. Efficient production of anthocyanins in Saccharomyces cerevisiae by introducing anthocyanin transporter and knocking out endogenous degrading enzymes. Front Bioeng Biotechnol. 2022;10:899182. https://doi.org/10.3389/fbioe.2022.899182.
Belwal T, Singh G, Jeandet P, Pandey A, Giri L, Ramola S, Bhatt ID, Venskutonis PR, Georgiev MI, Clément C, Luo Z. Anthocyanins, multi-functional natural products of industrial relevance: recent biotechnological advances. Biotechnol Adv. 2020;43:107600. https://doi.org/10.1016/j.biotechadv.2020.107600.
Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 2008;54:733–49. https://doi.org/10.1111/j.1365-313X.2008.03447.x.
Lu S, Zhuge Y, Hao T, Liu Z, Zhang M, Fang J. Systematic analysis reveals O-methyltransferase gene family members involved in flavonoid biosynthesis in grape. Plant Physiol Biochem. 2022;173:33–45. https://doi.org/10.1016/j.plaphy.2022.01.007.
Ueda M, Tanaka A. Genetic immobilization of proteins on the yeast cell surface. Biotechnol Adv. 2000;18:121–40. https://doi.org/10.1016/s0734-9750(00)00031-8.
Koopman F, Beekwilder J, Crimi B, van Houwelingen A, Hall RD, Bosch D, van Maris AJ, Pronk JT, Daran JM. De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb Cell Fact. 2012;11:155. https://doi.org/10.1186/1475-2859-11-155.
Levisson M, Patinios C, Hein S, de Groot PA, Daran JM, Hall RD, Martens S, Beekwilder J. Engineering de novo anthocyanin production in Saccharomyces cerevisiae. Microb Cell Fact. 2018;7:103. https://doi.org/10.1186/s12934-018-0951-6.
Lyu X, Zhao G, Ng KR, Mark R, Chen WN. Metabolic Engineering of Saccharomyces cerevisiae for De Novo production of Kaempferol. J Agric Food Chem. 2019;67:5596–606. https://doi.org/10.1021/acs.jafc.9b01329.
Du Y, Yang B, Yi Z, Hu L, Li M, Engineering. Saccharomyces cerevisiae Coculture Platform for the Production of Flavonoids. Journal of Agricultural and Food Chemistry. 2020;68:2146–2154. https://doi.org/10.1021/acs.jafc.9b07916
Guo Y, Dong J, Zhou T, Auxillos J, Li T, Zhang W, Wang L, Shen Y, Luo Y, Zheng Y, et al. YeastFab: the design and construction of standard biological parts for metabolic engineering in Saccharomyces cerevisiae. Nucleic Acids Res. 2015;43:e88. https://doi.org/10.1093/nar/gkv464.
Tian Y, Liu N, Zhao X, Mei X, Zhang L, Huang J, et al. Construction of anthocyanin biosynthesis system using chalcone as a substrate in Lactococcus lactis NZ9000. J Basic Microbiol. 2024:e2400274. https://doi.org/10.1002/jobm.202400274. Epub ahead of print.
Chen P, Zhang L, Chang N, Shi P, Gao T, Zhang L, Huang J. Preparation of virus-like particles for porcine circovirus type 2 by YeastFab Assembly. Virus Genes. 2018;54:246–55. https://doi.org/10.1007/s11262-018-1537-4.
Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. https://doi.org/10.1016/s0076-6879(02)50957-5.
Lõoke M, Kristjuhan K, Kristjuhan A. Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques. 2011;50:325–8. https://doi.org/10.2144/000113672.
Li J, Zhu K, Zhao H. Transcriptome analysis reveals the protection mechanism of proanthocyanidins for Saccharomyces cerevisiae during wine fermentation. Sci Rep. 2020;10:6676. https://doi.org/10.1038/s41598-020-63631-2.
Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE. 2008;3:e3647. https://doi.org/10.1371/journal.pone.0003647.
Zhang Y, Hu W, Peng X, Sun B, Wang X, Tang H. Characterization of anthocyanin and proanthocyanidin biosynthesis in two strawberry genotypes during fruit development in response to different light qualities. J Photochem Photobiol B. 2018;186:225–31. https://doi.org/10.1016/j.jphotobiol.2018.07.024.
Acknowledgements
We thank Prof. Junbiao Dai of Tsinghua University for providing us the YeastFab-related materials and methods.
Funding
The Innovation Center for Synthetic Biotechnology in China (TSBICIP-KJGG-014).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: DP H and JH H. Performed the experiments: XF M, DP H, N L, XW Z, TY J, BP Z. Analyzed the data: XF M, DP H, and N L. Contributed reagents/materials: LL Z, JH H, L Z. Wrote the paper: XF M and DP H.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mei, X., Hua, D., Liu, N. et al. De novo biosynthesis of anthocyanins in Saccharomyces cerevisiae using metabolic pathway synthases from blueberry. Microb Cell Fact 23, 228 (2024). https://doi.org/10.1186/s12934-024-02500-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-024-02500-3