Abstract
Background
Coronary heart disease (CHD) is a major global health concern, especially among individuals with type 2 diabetes (T2D). Given the crucial role of proteins in various biological processes, this study aimed to elucidate the aetiological role and predictive performance of protein biomarkers on incident CHD in individuals with and without T2D.
Methods
The discovery cohort included 1492 participants from the Cooperative Health Research in the Region of Augsburg (KORA) S4 study with 147 incident CHD cases (45 vs. 102 cases in the group with T2D and without T2D, respectively) during 15.6 years of follow-up. The validation cohort included 888 participants from the KORA-Age1 study with 70 incident CHD cases (19 vs. 51 cases in the group with T2D and without T2D, respectively) during 6.9 years of follow-up. We measured 233 plasma proteins related to cardiovascular disease and inflammation using proximity extension assay technology. Associations of proteins with incident CHD were assessed using Cox regression and Mendelian randomization (MR) analysis. Predictive models were developed using priority-Lasso and were evaluated on top of Framingham risk score variables using the C-index, category-free net reclassification index (cfNRI), and relative integrated discrimination improvement (IDI).
Results
We identified two proteins associated with incident CHD in individuals with and 29 in those without baseline T2D, respectively. Six of these proteins are novel candidates for incident CHD. MR suggested a potential causal role for hepatocyte growth factor in CHD development. The developed four-protein-enriched model for individuals with baseline T2D (ΔC-index: 0.017; cfNRI: 0.253; IDI: 0.051) and the 12-protein-enriched model for individuals without baseline T2D (ΔC-index: 0.054; cfNRI: 0.462; IDI: 0.024) consistently improved CHD prediction in the discovery cohort, while in the validation cohort, significant improvements were only observed for selected performance measures (with T2D: cfNRI: 0.633; without T2D: ΔC-index: 0.038; cfNRI: 0.465).
Conclusions
This study identified novel protein biomarkers associated with incident CHD in individuals with and without T2D and reaffirmed previously reported protein candidates. These findings enhance our understanding of CHD pathophysiology and provide potential targets for prevention and treatment.
Similar content being viewed by others
Background
Globally, coronary heart disease (CHD) is the leading cause of morbidity and mortality, particularly in Europe, where it accounts for nearly half of all deaths [1]. Although CHD incidence has declined in many countries in recent years, it continues to be a significant public health challenge. Type 2 diabetes (T2D) has been linked to an early onset of CHD and in middle-aged adults the risk of developing CHD is 2–4 times greater in persons with T2D than in those without T2D [2]. Moreover, established classical risk factors for CHD such as blood pressure, serum cholesterol, and smoking are more strongly associated with CHD in persons with diabetes than in those without [3, 4]. Thus, for the effective prevention and management of incident CHD, it is crucial to understand the underlying mechanisms leading to CHD in persons with and without diabetes in the general population.
Advanced proteomics methods such as proximity extension assay (PEA) technology allow the simultaneous measurement of hundreds and even thousands of protein biomarkers [5], which can contribute to the elucidation of unknown biochemical activities and pathways related to disease development and progression. Although several proteomics studies have been conducted for incident CHD [6,7,8,9,10], only a few biomarkers are considered as reliable predictors in clinical practice and treatment guidelines [1, 11] and studies stratifying by diabetes status are lacking. As our and other studies have previously shown, prevalent T2D is strongly associated with various protein biomarkers [12,13,14,15,16]. Furthermore, Elhadad et al. conducted a bidirectional Mendelian randomization (MR) analysis, providing further evidence regarding the influence of T2D on protein levels [13]. Thus, it seems likely that protein–CHD associations could be affected by diabetes status.
Hence, the present study, conducted in the Cooperative Health Research in the Region of Augsburg (KORA) S4 cohort with a 16-year follow-up, explored the potential associations between protein biomarkers and incident CHD separately in individuals with and without T2D. This endeavor aimed to identify both unique and shared pathophysiological pathways and biomarkers potentially involved in the development of CHD in different diabetes states. In addition, we performed MR analysis to further elucidate possible causal effects of the identified biomarkers on incident CHD. Lastly, we evaluated if the identified protein biomarkers improved the predictive performance of incident CHD on top of traditional risk factors for CHD [17]. Our findings were subsequently validated in the prospective KORA-Age1 cohort study among older participants from the general population followed for up to 7.6 years.
Methods
Study population
The discovery sample was derived from the population-based KORA S4 cohort study comprising 4261 participants at baseline (1999 to 2001) [18]. The present analysis was confined to individuals aged 55–74 years due to the availability of proteomics data, resulting in a sample of 1653 participants who were followed for a median duration of 15.6 years. After exclusion of participants with missing proteomics data and those with non-T2D (type 1 diabetes and drug-induced diabetes), unclear diabetes status, missing covariables of the main model in the association analysis, prevalent CHD, and those lost to follow-up, a total of 1492 participants remained for analysis (see Supplementary Fig. 1, Additional file 1). Prevalent T2D comprised persons with self-reported and subsequently validated clinically diagnosed T2D and persons with newly diagnosed T2D based on an oral glucose tolerance test (OGTT) using the WHO criteria [19] or baseline glycated hemoglobin (HbA1c) levels ≥ 6.5%. Self-reported T2D was confirmed through questionnaires sent to treating physicians or through medical record reviews. Participants were classified as having clinically diagnosed T2D only if the treating physician reported a diagnosis of T2D, if T2D was documented in the medical records, or if the participants reported taking antidiabetic medication. Finally, the discovery study comprised 228 participants with T2D and 1264 participants without T2D.
For validation, data from the KORA-Age1 study was used. This study includes all participants of the four cross-sectional Monitoring of Trends and Determinants in Cardiovascular Disease (MONICA) Augsburg / KORA surveys conducted in 1984/85 (Survey S1), 1989/90 (Survey S2), 1994/95 (Survey S3), and 1999/2001 (Survey S4), who were born in 1943 or earlier, comprising 9197 participants [20]. Out of these, a sex- and age-stratified random sample of 1079 individuals was extensively examined in 2009 including the collection of blood samples. In the present analysis, data from these participants were used for the validation of the results of the KORA S4 study. After exclusions (see Supplementary Fig. 1, Additional file 1), 888 participants aged 65–93 years who were followed for a median duration of 6.9 years remained for analysis. In the validation study, prevalent T2D was defined based on self-report with subsequent validation as described above, and baseline HbA1c levels ≥ 6.5% only, since no OGTT was conducted in the KORA-Age1 study. Finally, the validation study included 165 participants with T2D and 723 participants without T2D. Out of the 888 participants of KORA-Age1, 206 participants are also part of the KORA S4 discovery sample, since the S4 participants falling into the respective age range were also invited to be part of KORA-Age1 at a later time point as described above.
Proteomics measurements
At the baseline examinations, venous blood samples were collected while sitting. Plasma samples were stored in liquid nitrogen at − 196 °C until proteomics analysis in 2019–2020 for KORA S4 and in 2023 for KORA-Age1.
The PEA technology by Olink® (Olink Proteomics, Uppsala, Sweden) was used to measure 276 EDTA plasma proteins related to cardiovascular diseases (CVD) and inflammation (CVD-II, CVD-III, and Inflammation panels) in both KORA S4 and KORA-Age1. Detailed measurement procedures were previously outlined [21]. Log2-normalized protein expression values were provided and were normalized by their respective standard deviations within the complete dataset before applying exclusions. Consistent quality control criteria were applied to both the KORA S4 and KORA-Age1 proteomics data. Proteins with over 25% of values below the limit of detection (LOD) were excluded, and proteins measured in duplicate were resolved by retaining the duplicate with fewer LOD values and a lower inter-assay coefficient of variation. Additionally, proteins with missing values were excluded. In the KORA S4 cohort, a total of 233 protein biomarkers were finally included. 76 identified biomarkers associated with incident CHD in the KORA S4 dataset were relevant for the KORA-Age1 validation analysis, and 75 of these biomarkers were included after quality control.
Outcomes
The combined outcome of CHD encompassed nonfatal myocardial infarction (MI), coronary death, and sudden death, as classified by the International Classification of Disease 9th Revision (410–414 and 798). Until December 2000, the diagnosis of major nonfatal MI was based on the MONICA study algorithm, which considered factors such as symptoms, cardiac enzyme levels (including creatine kinase, aspartate aminotransferase, and lactate dehydrogenase), 12-lead electrocardiograms (ECGs), autopsy results, and history of CHD in fatal cases [22]. From January 2001 onwards, the criteria for diagnosing MI followed the guidelines established by the European Society of Cardiology and the American College of Cardiology [23].
Cases of incident CHD were identified through the KORA Augsburg MI registry, which systematically tracked all fatal and nonfatal MI, in or out of hospital, among residents within the study region aged 25 to 84 years from 2009 onwards [24]. Additionally, regular follow-up questionnaires were administered to the participants. Self-reported incident cases occurring outside the study area and those with self‐reported date of diagnosis falling out of the age range that was covered by the MI registry were further validated using hospital records or by contacting the treating physician. Validation for all coronary deaths was performed through autopsy reports, death certificates, chart reviews or information from the last treating physician. During the study period, KORA S4 study participants underwent two follow-up examinations in 2006–2008 and 2013–2014, which included self-reported information on health status. To further enrich the dataset, postal questionnaires soliciting self-reported health details were dispatched to S4 participants in 2008–2009 and 2016. In contrast, KORA-Age1 participants experienced a singular follow-up examination in 2012 and received postal questionnaires in 2016.
Baseline measurements / covariates
All participants underwent standard physical and medical examinations at KORA S4 and KORA-Age1 [20, 25]. Trained medical staff conducted interviews to collect information on age, sex, education, smoking habits, alcohol consumption, physical activity, and medical history. Educational attainment was recorded as completed years of schooling. Smoking status was categorized as either current smoker or non-smoker (including never and former smokers). Alcohol intake was categorized into three groups: no consumption (0 g/day), moderate consumption (men: 0.1–39.9 g/day, women: 0.1–19.9 g/day), and high consumption (men: ≥40 g/day, women: ≥20 g/day), based on their self-reported consumption of beer, wine, and liquor on two weekdays and the weekend. Physical activity was assessed as either active or inactive, factoring in the frequency and duration of weekly exercise across different seasons. Medication usage, such as antihypertensive and lipid-lowering drugs, was defined using Anatomical Therapeutic Chemical Classification System codes. Enzymatic methods were used to measure total cholesterol and high-density lipoprotein cholesterol (HDL-cholesterol). Body mass index (BMI) was calculated as weight (kg) divided by height squared (m²). Systolic and diastolic blood pressure were measured on the right arm in a sitting position following the World Health Organization MONICA protocol [26]. Participants without diabetes received a standard 75 g OGTT test in KORA S4. Their blood samples to measure diabetes parameters were taken without stasis after an overnight fast of ≥ 8 h as well as 2 h after the glucose solution ingestion [18].
Statistical analysis
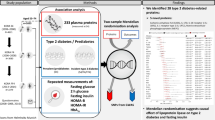
The analysis strategy of the study is shown in Fig. 1.
Association analyses of protein biomarkers with coronary heart disease
Associations between each plasma protein level and time to incident CHD were assessed for participants with and without T2D at baseline using Cox proportional hazard regression models. The association analysis was adjusted for important CHD risk factors at baseline: Model 1 (main model) was adjusted for age, sex, total cholesterol, HDL-C, systolic blood pressure, smoking status, and antihypertensive medication usage. These covariates, along with diabetes status, constitute the Framingham Risk Score (FRS) for CHD [17]. Model 2 was further adjusted for additional cardiovascular-related risk factors including BMI, education years, physical activity, alcohol consumption, and lipid-lowering medication usage. Model 3 included fasting status as an independent variable in addition to model 1. The interaction effect of diabetes status was examined by adding diabetes status and the term (protein×diabetes status) to model 1 among the whole KORA S4 participants. Nominally significant (p-value < 0.05) proteins in model 1 were validated in the KORA-Age1 study using the same model 1. Proteins with validated significance at the false discovery rate (FDR) lower than 0.05 (p_FDR < 0.05), calculated based on the number of nominally significant proteins in KORA S4 (4 for the group with T2D and 71 for the group without T2D), were selected.
To assess whether death as a competing risk influenced the validated associations, we performed a sensitivity analysis using the Fine-Gray subdistribution hazard model to estimate the CHD incidence over time in the presence of death as a competing risk. In another sensitivity analysis, we excluded the overlapping participants, who participated in both the KORA S4 and KORA-Age1 study, from the KORA-Age1 analysis (n = 37 in the group with T2D and n = 169 in the group without T2D).
Two-sample mendelian randomization analysis
A two-sample MR was applied using the published available genome-wide association study (GWAS) data of European ancestry to explore the potential causal links between biomarkers and incident CHD. For the validated protein biomarkers, the instrumental variables (IVs) were extracted from the Olink-based GWAS database, which included 54,219 participants from the UK biobank [27]. Incident CHD GWAS data were from Mbatchou J et al., involving 352,063 participants from the UK Biobank dataset (case-control ratio = 1:11) [28]. The IV selection involved identifying SNPs associated with proteins at a significance threshold of p-value < 5 × 10− 8, focusing on cis regions, and eliminating ambiguous palindromic SNPs with A/T or G/C alleles. To test the assumption of MR, SNP independence was verified via PhenoScanner V2 database (http://www.phenoscanner.medschl.cam.ac.uk/), and SNPs associated with traditional CHD risk factors were removed. The remaining SNPs were clumped using an r2 = 0.001 threshold to eliminate the linkage disequilibrium with the lead SNP. SNPs were then extracted as IVs from the outcome’s GWAS.
For causal assessment, the Wald ratio test was performed when only one IV was available, and the inverse variance-weighted method was used for proteins with at least two IVs [29, 30]. The significant threshold was adjusted using the Bonferroni correction (p-value = 0.05 divided by the number of tested proteins). Sensitivity analyses evaluated instrument heterogeneity and directional horizontal pleiotropy using Cochran’s Q test and MR-Egger regression.
Network analysis and enrichment analysis
To elucidate potential connections and mechanisms of the selected proteins, we annotated the above validated two proteins in the group with T2D and 29 proteins in the group without T2D, respectively, using the STRING database version 12.0 (https://string-db.org/). Based on the built network of identified proteins, enrichment analysis was performed to detect pathways linked to CHD based on the Reactome pathway knowledgebase [31]. Given the limited pool of identified proteins (n = 2) in the group with T2D, the enrichment analysis was performed only for the 29 proteins identified in the group without T2D.
Prediction of incident coronary heart disease
KORA S4 served as the discovery dataset, while KORA-Age1 was used as the validation dataset in the prediction analysis. The components of the FRS (model 1 in the association analyses) were used as the basic model for CHD prediction.
To enhance the accuracy and effectiveness of constructing a predictive model, only the protein biomarkers significantly associated with incident CHD in the discovery analysis were included in the predictor selection for the extended model (basic model + protein biomarkers) using the priority-Lasso which is a least absolute shrinkage and selection operator (LASSO)-based intuitive analysis strategy and constructs a prediction model for a clinical outcome by defining the blocks of different types of predictor variables [32]. There were five and 72 proteins selected for participants with T2D and without T2D in KORA S4, respectively, of which one in both groups (melusin [ITGB1BP2]) failed quality control in KORA-Age1 and was therefore excluded from further analyses. The penalization parameter λ was determined by five-fold cross-validation with Cox regression design. In the discovery dataset, we fixed the seven FRS variables as block 1 to prevent any shrinkage by priority-Lasso, while the identified proteins (4 / 71 proteins) were incorporated as block 2 for their respective T2D status groups. The performance of the priority-Lasso protein-extended model was compared to the basic model in both the KORA S4 dataset and the KORA-Age1 dataset. The performance of the basic and extended model was evaluated through three measures: (1) Harrel’s concordance index (C-index) for the basic model, the protein-extended model, and their difference (ΔC‐index = C‐index extended - C‐index basic) [33]; (2) the category-free net reclassification index for all participants combined (cfNRI), for incident CHD cases (cfNRIcases), and for non-CHD controls (cfNRIcontrols) [34]; (3) the absolute integrated discrimination improvement (IDI) [35]. All effect estimates were calculated as the arithmetic mean of these measures using five-fold cross-validation. Their corresponding confidence intervals were calculated using 100x bootstrapping.
The R version 4.3 (https://www.r-project.org/) was used for all analyses.
Results
Baseline characteristics of the study participants
Table 1 presents the characteristics of the study participants at baseline. In the KORA S4 study, 45 and 102 participants had incident CHD in the group with and without T2D at baseline (15.5 vs. 5.6 per 1000 person-years), respectively. In the KORA-Age1 study, 19 and 51 participants had incident CHD in the group with and without T2D at baseline (18.7 vs. 10.7 per 1000 person-years), respectively.
Associations of protein biomarkers with coronary heart disease
In the KORA S4 study, five protein biomarkers showed nominally significant associations with incident CHD in the group with T2D, whereas a total of 72 biomarkers were significant in the group without T2D in model 1 (see Supplementary Table 1, Additional file 2). ITGB1BP2 failed the quality control in KORA-Age1 and was consequently excluded from the validation study. Of the remaining 4 and 71 biomarkers, two and 29 protein biomarkers, respectively, were successfully validated in the KORA-Age1 dataset after correction for multiple testing (see Fig. 2 and Supplementary Table 2, Additional file 2). The correlations between these 31 validated protein biomarkers are illustrated in Supplementary Fig. 2, Additional file 1. Notably, there was no overlap in significant proteins between the two distinct diabetes status groups in the validation study.
After further adjusting for other lifestyle factors in model 2, five of the validated proteins (osteoclast-associated immunoglobulin-like receptor [HOSCAR], placenta growth factor [PGF], thrombospondin-2 [THBS2], ST2 protein, and tumor necrosis factor receptor 1 [TNF-R1]) lost significance in the group without T2D. After further accounting for baseline fasting status in model 3, all associations from model 1 remained significant (see Supplementary Table 1, Additional file 2). In the KORA S4 study population which was used as the discovery study, 51 proteins showed significant interactions with T2D status supporting our hypothesis that associations of proteins with CHD may be modified by the presence of T2D. Eight out of the 33 validated proteins displayed a significant interaction effect with diabetes status in the discovery study (see Supplementary Table 1, Additional file 2), but none of these interaction effects were validated in the KORA-Age1 study.
Considering death as a competing risk, two of the validated proteins (tumor necrosis factor receptor superfamily member 9 [TNFRSF9], and fatty acid-binding protein 4 [FABP4]) were no longer significantly associated with incident CHD in both cohorts in those without T2D (see Supplementary Table 3, Additional file 2). After excluding overlapping KORA S4 participants from the KORA-Age1 sample, eight proteins (C-X-C motif chemokine 9 [CXCL9], interleukin-2 receptor subunit alpha [IL-2RA], follistatin [FS], matrix metalloproteinase-12 [MMP-12], hepatocyte growth factor [HGF], oncostatin-M [OSM], TNFRSF9, and scavenger receptor cysteine-rich type 1 protein M130 [CD163]) lost significance after multiple testing regarding their association with incident CHD (see Supplementary Table 4, Additional file 2).
Association of 233 proteins with incident coronary heart disease in individuals (a) with type 2 diabetes (T2D), and (b) without T2D at baseline. Hazard ratios have been calculated per 1 SD increase in normalized protein expression values on a log2 scale. Effect estimates and p-values were derived from Cox regression analysis adjusted for age, sex, total cholesterol, high-density lipoprotein cholesterol, systolic blood pressure, antihypertensive medication use, and current smoking (Model 1). The red triangles represent the validated proteins in the validation study, identified using the false discovery rate (p_FDR < 0.05). The black dots represent significant proteins at the uncorrected level (p < 0.05) in the discovery study which were not replicated in the validation study. (c) Forest plot of validated proteins in KORA S4 and KORA-Age1 cohorts stratified by T2D status
Causal effects of validated proteins on coronary heart disease
Cis-acting genetic IVs were identified for 29 validated CHD-associated proteins from previous GWAS data and their potential causal effects were assessed (see Fig. 3 and Supplementary Table 5, Additional file 2). Two proteins (eukaryotic translation initiation factor 4E-binding protein 1 [4E-BP1] and CD163) lacked qualified IVs. HGF was the only protein with a statistically significant causal effect on CHD after correction for multiple testing (Wald ratio, b = 0.3422; p-value = 0.0004), while PGF lost significance following correction for multiple testing (Wald ratio, b = 0.1607; p-value = 0.0068).
Network and related pathways of identified proteins
To unravel biological insights, network analysis and pathway analysis were performed on the 29 validated proteins in the group without T2D at baseline. The following Reactome pathways (number of involved proteins) were overrepresented in the protein biomarker set: immune system (n = 15), cytokine signaling in immune system (n = 11), signaling by interleukins (n = 8), and TNFR2 non-canonical NF-kB pathway (n = 4) and PI5P, PP2A and IER3 regulate PI3K/AKT signaling (n = 4). The resulting protein-protein interaction network is visualized in Fig. 4. TNF-R1 was involved in the top four pathways and emerged as a central player within the network.
Protein-protein interaction network of validated coronary heart disease-associated proteins among participants without type 2 diabetes at baseline. The edges between protein nodes represent the interaction score between the proteins from the STRING database considering all types of evidence. Only edges featuring interaction scores > 0.15 are displayed. The thickness of edges corresponds to the strength of data support. Node color signifies the Reactome pathway the protein is associated with. The 5 most enriched Reactome pathways are displayed
Prediction of incident coronary heart disease
In the group with T2D, the four identified proteins (carboxypeptidase A1 [CPA1], IL-2RA, CXCL9, and neurotrophin-3 [NT-3]) were all selected by priority-Lasso on top of the basic model. While the basic model yielded a C-index of 0.693 (95% CI = 0.563–0.774), the protein-extended model showed improved predictive performances with ΔC-index of 0.017 (95% CI = 0.006–0.145), cfNRI of 0.253 (95% CI = 0.024–0.497) and IDI of 0.051 (95% CI = 0.014–0.124) in the KORA S4 sample. In the KORA-Age1 sample, the protein-extended model improved only the cfNRI with a value of 0.633 (95% CI = 0.139–1.075) which was mainly driven by an increased cfNRIcontrols (0.506, 95% CI = 0.288–0.796).
In the group without T2D, 12 proteins (CD163, epithelial cell adhesion molecule [Ep-CAM], osteopontin [OPN], TNF-R1, kidney injury molecule 1 [KIM1], proheparin-binding EGF-like growth factor [HB-EGF], MMP-12, protein-glutamine gamma-glutamyltransferase 2 [TGM2], vascular endothelial growth factor A [VEGF-A], interleukin-6 [IL-6], IL-10, and protein S100-A12 [EN-RAGE]) were selected alongside the basic FRS variables in the priority-Lasso analysis. The basic model yielded a C-index of 0.700 (95% CI = 0.657–0.760) in the KORA S4 sample and 0.683 (95% CI = 0.564–0.769) in the KORA-Age1 sample, respectively. This augmented model led to improved predictive performances in both the KORA S4 and KORA-Age1 datasets, yielding enhanced ΔC-index (0.038, 95% CI = 0.024–0.133) and overall cfNRI (0.465, 95% CI = 0.027–0.741) in the KORA-Age1 sample which was mainly driven by an increased cfNRIcontrols (0.380, 95% CI = 0.273–0.533) as depicted in Table 2. The estimates of the extended model for incident CHD in KORA S4 are presented in Supplementary Tables 6–7, Additional file 2.
In the sensitivity analysis excluding 206 overlapping participants from the KORA-Age1 sample, prediction results were very similar. In the group with T2D, the established model showed improved predictive performances based on cfNRI (0.522 [95% CI = 0.363–0.925]) which was mainly driven by the cfNRIcontrols (0.488 [95% CI = 0.329–0.625]), while in the group without T2D, an enhanced ΔC-index of 0.048 (95% CI = 0.029–0.130) and an overall cfNRI of 0.262 (95% CI = 0.003–0.760) were observed (see Supplementary Table 8, Additional file 2).
Discussion
We conducted a longitudinal analysis to investigate the proteomic profile of incident CHD among individuals with different baseline diabetes status. Only two validated proteins were identified for incident CHD in individuals with T2D, while twenty-nine validated proteins were identified in those without T2D, respectively. Among the 31 proteins, six proteins (TNFRSF13B, THBS2, transforming growth factor-alpha [TGF-alpha]), CXCL9, CXCL11, and 4E-BP1) are novel candidate biomarkers for CHD. Additionally, the two-sample MR approach provided suggestive evidence for a causal effect of HGF on CHD.
Novel protein biomarkers associated with CHD
Several novel incident CHD-related protein biomarkers identified in the present study have previously been demonstrated to be related to atherosclerosis and CHD progression. Among these, increased serum levels of TNFRSF13B, a TNF superfamily receptor, have been linked to the presence of plaque, i.e. subclinical CHD [36]. Furthermore, we identified THBS2 as a marker of incident CHD, which is a matricellular protein facilitating cell-matrix interactions, that was positively associated with both incident heart failure (HF) hospitalization and deterioration in diastolic function in a recent study [37]. TGF-alpha, which directly activates the transcription factor NF-κB through the epidermal growth factor receptor pathway, was previously found to be associated with higher cardiovascular mortality in patients with chronic CHD [38].
Other novel biomarkers identified by our study include CXCL9 and CXCL11, which are inflammatory chemokines known to induce immune cell infiltration through the C-X-C motif chemokine receptor 3 (CXCR3). Previous investigations have suggested the involvement of CXCLs and CXC receptors in distal sensorimotor polyneuropathy, various CVDs as well as T2D [39,40,41]. This may partly elucidate the specific role of CXCL9, particularly in individuals with baseline T2D in the context of incident CHD. Furthermore, we found a positive association between 4E-BP1 and incident CHD, which is a substrate of the mTOR-containing multiprotein complex-1 (mTORC1) with the capacity to inhibit translation initiation. This protein has been assumed to play a crucial role in regulating the viability of cardiomyocytes, particularly in the context of heart failure [42]. This novel set of protein biomarkers presents new avenues for exploring potential prevention strategies and therapeutic targets addressing CHD.
Confirmed protein biomarkers associated with CHD
Our validated CHD-related biomarkers align with previous investigations of proteomic biomarkers using the same Olink panels to identify associations with incident CHD [6,7,8,9, 43, 44]. It is noteworthy that the aforementioned studies predominantly assessed associations of proteins with incident CHD in population-based samples comprising participants with and without diabetes together, whereas our study stands out as the first to identify biomarkers specifically according to baseline T2D status. Growth differentiation factor-15 (GDF-15), a member of the transforming growth factor-β cytokine superfamily, is known to severely increase during oxidative stress and inflammation, which suggests GDF-15 as a credible marker for the increased risk of incident CHD [6, 9]. TNF-related apoptosis inducing receptor 2, a TNF superfamily member, has been associated with a higher risk of incident MI, possibly due to its role in inflammation and apoptosis [6]. One of the metalloproteases involved in the breakdown of collagen and elastin, MMP-12, has also been positively associated with both incident MI and HF [6, 9]. Another of the identified proteins that plays a role in inflammation is urokinase plasminogen activator surface receptor, which is closely linked with immune and inflammatory activation and was associated with an increasing risk of incident MI [6]. FABP4, which is secreted by adipocytes, has well-documented implications for insulin resistance and atherosclerosis, consistently showing elevated levels in persons developing incident CHD [7]. Similar results in studies on incident CHD and cardiovascular mortality have been reported for CD163, a marker involved in macrophage activation [7, 8]. Additionally, FS is secreted from the liver and was reported to be associated with a higher risk of incident coronary events, independently of established risk factors including diabetes, using PEA technology [43]. Cystatin B, an inhibitor of cathepsin L, was associated with an elevated risk of incident CHD in the highest tertile [44].
Similar findings for incident CHD were observed regarding HGF, TNF-R1, PGF, EN-RAGE, and IL-2RA when proteins were measured using other methods, such as ELISAs. HGF in particular, emerges as a pivotal protein known for its effects on CVD, activating pathways that counteract apoptosis, inflammation, oxidation, and fibrosis [45]. Our MR analysis revealed a suggestive positive causal association of HGF on incident CHD in the general population, with a consistent directionality with our findings of association analysis in individuals without baseline T2D. Previous study supported our observational and MR analysis findings [46]. Of note, this is the first study that provides suggestive evidence for a causal association between HGF and incident CHD. TNF-R1, a crucial proinflammatory cytokine mediator, was associated with an increased risk of incident CHD, especially in women [47]. PGF, a VEGF homologue and EN-RAGE, an endogenously produced inflammatory ligand, were associated with a higher risk of incident CHD [48, 49]. Moreover, serum IL-2RA, a marker of T lymphocyte activation, was significantly positively associated with incident CHD in participants with T2D in the present study. In line with our findings, an increased risk for incident CHD as well as prevalent T2D was reported in older adults, but the effect of IL-2RA was not specifically tested on incident CHD in persons with baseline T2D [50].
In line with our findings, cathepsin L1, gal-9, spondin-2, and TNFRSF11A measured using PEA technology were significantly altered in patients with prevalent CHD compared to participants without CHD [51]. Additionally, other proteins including HB-EGF, IL-27, sortilin, matrix metalloproteinase-1, OSM, ST2, TNFRSF9, and HOSCAR were reported to show higher concentrations in blood samples from individuals with CHD compared with healthy controls or non-CHD participants [52,53,54,55,56,57,58,59].
Our pathway and network analysis for the 29-incident CHD-related protein biomarkers among individuals without baseline T2D revealed insights into the mechanistic underpinnings of CHD pathogenesis. Notably, the enrichment of pathways such as the immune system, cytokine signaling in the immune system, signaling by interleukins, and the TNFR2 non-canonical NF-kB pathway underscored the role of inflammatory processes in CHD development. This aligned with established literature highlighting the significance of immune response and cytokine signaling in the development of CHD [60, 61]. Importantly, TNF-R1, a central player in our identified pathways, has been implicated in mediating inflammatory responses, reinforcing its potential key role in the pathogenesis of incident CHD. Our findings exhibited substantial overlap with previously identified pathways [60, 61], providing further support for the involvement of inflammatory mechanisms in CHD development.
Prediction of CHD through protein biomarkers
Our study is the first to establish proteomics-enriched predictive models for incident CHD separately for those with and without prevalent T2D. However, it is noteworthy that in the validation study, protein-enriched models significantly improved the predictive performance based on selected performance measures only, particularly among those with T2D. Among participants without T2D, the model enriched with 12 proteins improved discrimination of incident CHD by 5.6% based on the C-index (delta C-index = 0.038) compared with traditional CHD risk factors in the validation study. Our findings partly coincide with those of Lind et al., who utilized the Olink CVD I panels to derive a 7-protein enriched model for the prediction of the 15-year risk of incident CVD (including MI, ischemic stroke and HF) [6] in a population including about 11% of persons with prevalent diabetes. This approach resulted in a 7.3% improvement compared with traditional risk factors in the replication sample. Hereby, EN-RAGE was the only biomarker that overlapped with our selected proteins. Similarly, McCarthy et al. established a protein model measured using the Luminex xMAP platform and reported a 3.6% improvement in predicting incident major adverse cardiovascular events (including cardiovascular death, MI, and stroke) during a 3.6-year follow-up period [62]. However, the application of these models in clinical practice needs careful consideration, given the differences in biomarkers, populations, and methodologies across studies.
Study strengths and limitations
We used advanced targeted proteomics technology to examine a wide range of proteins linked to CHD. A major strength of the statistical analysis constitutes the validation of the identified proteins in another cohort study. Specifically exploring protein-CHD associations by diabetes status provided evidence of the underlying mechanisms leading to CHD in persons with and without T2D. By analyzing genetic data using a Mendelian randomization approach, we gained insights into potential causal relationships between proteins and CHD risk.
However, there are some limitations to consider. First, due to the limited number of incident CHD cases, our analyses may not be sufficiently powered to detect a difference in CHD vs. no CHD groups, particularly in the group with T2D at baseline. Along these lines, due to the limited number of validated proteins in those with T2D, pathway analyses had to be restricted to those without T2D. Adjusting for multiple testing was necessary due to the numerous analyses, but could have caused overcorrection. Additionally, we lacked OGTT data to identify previously unknown diabetes in the validation cohort. However, the differences in effect estimates were relatively small when shifting those with newly diagnosed diabetes from the group with diabetes to the group without diabetes in KORA S4 (see Supplementary Table 9, Additional file 2). While validation in the KORA-Age1 cohort strengthens the results for the validated proteins, we may have lacked replication for some proteins particularly if their impact was modified by age since the KORA-Age1 study participants were all older than 65 years. It is worth noting that there is some overlap between the participants in KORA-Age1 and KORA S4 cohort. However, these overlapping participants were examined twice at different time points. Importantly, when we excluded these overlapping participants from our analyses, the results did not show substantial changes. In addition, the shorter follow-up duration in KORA-Age1 (median follow-up time: 6.9 years) compared to KORA S4 (median follow-up time: 15.6 years) should be acknowledged as a limitation in interpreting our findings. To ensure broader applicability, further validation across diverse age groups, ethnicities, and regions are necessary. Moreover, due to the limitations of the GWAS database of incident CHD, the MR analysis performed in this study verified the causal impact of protein biomarkers in the general population rather than in populations with different baseline diabetes status. Additionally, the practical value of the identified proteomic markers for predicting CHD risk needs to be tested in larger studies covering a wider age range.
Conclusions
In summary, we identified two and 29 validated protein candidates possibly involved in the pathophysiology of CHD among individuals with and without baseline T2D, respectively. Our results provide new insights into a possible causal role of plasma HGF on CHD development and additional support for the involvement of inflammatory processes in CHD development particularly among those without T2D at baseline. Moreover, we established a protein-enriched CHD risk factor-based model which improved the predictive performance of incident CHD in persons with or without T2D compared to the traditional CHD risk factor model. Further research examining larger numbers of T2D patients will be crucial to verify the importance of specific pathways in those with T2D.
Data availability
The datasets from this KORA study are not publicly available because the datasets are subject to national data protection laws, and restrictions were imposed by the ethics committee of the Bavarian Chamber of Physicians to ensure data privacy of the study participants. However, datasets are available from the corresponding author on reasonable request through a project agreement from KORA (https://helmholtz-muenchen.managed-otrs.com/external/). Requests should be sent to kora.passt@helmholtz-munich.de and are subject to approval by the KORA board.
Abbreviations
- 4E-BP1:
-
Eukaryotic translation initiation factor 4E-binding protein 1
- BMI:
-
Body mass index
- CD163:
-
Scavenger receptor cysteine-rich type 1 protein M130
- cfNRI:
-
Category-free net reclassification index
- CHD:
-
Coronary heart disease
- C-index:
-
Harrel’s concordance index
- CPA1:
-
Carboxypeptidase A1
- CVD:
-
Cardiovascular disease
- CXCL:
-
C-X-C motif chemokine
- EN-RAGE:
-
Protein S100-A12
- Ep-CAM:
-
Epithelial cell adhesion molecule
- FABP4:
-
Fatty acid-binding protein 4
- FDR:
-
False discovery rate
- FRS:
-
Framingham Risk Score
- FS:
-
Follistatin
- GDF-15:
-
Growth differentiation factor 15
- GWAS:
-
Available genome-wide association study
- HbA1c:
-
Glycated hemoglobin
- HB-EGF:
-
Proheparin-binding EGF-like growth factor
- HDL-cholesterol:
-
High-density lipoprotein cholesterol
- HF:
-
Heart failure
- HGF:
-
Hepatocyte growth factor
- HOSCAR:
-
Osteoclast-associated immunoglobulin-like receptor
- IDI:
-
Integrated discrimination improvement
- IL:
-
Interleukin
- IL-2RA:
-
Interleukin-2 receptor subunit alpha
- IV:
-
Instrumental variable
- KIM1:
-
Kidney injury molecule 1
- KORA:
-
Cooperative Health Research in the Region of Augsburg
- LASSO:
-
Least absolute shrinkage and selection operator
- LOD:
-
Limit of detection
- ITGB1BP2:
-
Melusin
- MI:
-
Myocardial infarction
- MMP-12:
-
Matrix metalloproteinase-12
- MONICA:
-
Monitoring of Trends and Determinants in Cardiovascular Disease
- MR:
-
Mendelian randomization
- NT-3:
-
Neurotrophin-3
- OGTT:
-
Oral glucose tolerance test
- OPN:
-
Osteopontin
- OSM:
-
Oncostatin-M
- PEA:
-
Proximity extension assay
- PGF:
-
Placenta growth factor
- T2D:
-
Type 2 diabetes
- TGF-alpha:
-
Transforming growth factor-alpha
- THBS2:
-
Thrombospondin 2
- TNF-R1:
-
Tumor necrosis factor receptor 1
- TGM2:
-
Protein-glutamine gamma-glutamyltransferase 2
- TNFRSF:
-
Tumor necrosis factor receptor superfamily member
- VEGF-A:
-
Vascular endothelial growth factor A
References
Visseren FLJ, Mach F, Smulders YM, Carballo D, Koskinas KC, et al. 2021 ESC guidelines on cardiovascular disease prevention in clinical practice. Eur Heart J. 2021;42(34):3227–337.
Sattar N, Rawshani A, Franzen S, Rawshani A, Svensson AM, et al. Age at diagnosis of type 2 diabetes Mellitus and associations with Cardiovascular and Mortality risks. Circulation. 2019;139(19):2228–37.
Fuller JH, Shipley MJ, Rose G, Jarrett RJ, Keen H. Mortality from coronary heart disease and stroke in relation to degree of glycaemia: the Whitehall study. Br Med J (Clin Res Ed). 1983;287(6396):867–70.
Rosengren A, Welin L, Tsipogianni A, Wilhelmsen L. Impact of cardiovascular risk factors on coronary heart disease and mortality among middle aged diabetic men: a general population study. BMJ. 1989;299(6708):1127–31.
Wik L, Nordberg N, Broberg J, Bjorkesten J, Assarsson E, et al. Proximity extension assay in combination with next-generation sequencing for high-throughput proteome-wide analysis. Mol Cell Proteomics. 2021;20:100168.
Lind L, Arnlov J, Sundstrom J. Plasma protein Profile of Incident Myocardial Infarction, ischemic stroke, and Heart failure in 2 cohorts. J Am Heart Assoc. 2021;10(12):e017900.
Molvin J, Jujic A, Melander O, Pareek M, Rastam L, et al. Proteomic exploration of common pathophysiological pathways in diabetes and cardiovascular disease. ESC Heart Fail. 2020;7(6):4151–8.
Ong KL, Chung RWS, Hui N, Festin K, Lundberg AK, et al. Usefulness of certain protein biomarkers for prediction of Coronary Heart Disease. Am J Cardiol. 2020;125(4):542–8.
Lind L, Gigante B, Borne Y, Malarstig A, Sundstrom J, et al. The plasma protein profile and cardiovascular risk differ between intima-media thickness of the common carotid artery and the bulb: a meta-analysis and a longitudinal evaluation. Atherosclerosis. 2020;295:25–30.
Ferreira JP, Sharma A, Mehta C, Bakris G, Rossignol P, et al. Multi-proteomic approach to predict specific cardiovascular events in patients with diabetes and myocardial infarction: findings from the EXAMINE trial. Clin Res Cardiol. 2021;110(7):1006–19.
Goff DC Jr., Lloyd-Jones DM, Bennett G, Coady S, D’Agostino RB, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice guidelines. Circulation. 2014;129(25 Suppl 2):49–73.
Beijer K, Nowak C, Sundstrom J, Arnlov J, Fall T, et al. In search of causal pathways in diabetes: a study using proteomics and genotyping data from a cross-sectional study. Diabetologia. 2019;62(11):1998–2006.
Elhadad MA, Jonasson C, Huth C, Wilson R, Gieger C, et al. Deciphering the plasma proteome of type 2 diabetes. Diabetes. 2020;69(12):2766–78.
Gudmundsdottir V, Zaghlool SB, Emilsson V, Aspelund T, Ilkov M, et al. Circulating protein signatures and causal candidates for type 2 diabetes. Diabetes. 2020;69(8):1843–53.
Ferreira JP, Pizard A, Machu JL, Bresso E, Rocca HB, et al. Plasma protein biomarkers and their association with mutually exclusive cardiovascular phenotypes: the FIBRO-TARGETS case-control analyses. Clin Res Cardiol. 2020;109(1):22–33.
Luo H, Bauer A, Nano J, Petrera A, Rathmann W, et al. Associations of plasma proteomics with type 2 diabetes and related traits: results from the longitudinal KORA S4/F4/FF4 study. Diabetologia. 2023;66(9):1655–68.
D’Agostino RB, Sr., Vasan RS, Pencina MJ, Wolf PA, Cobain M, et al. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation. 2008;117(6):743–53.
Rathmann W, Haastert B, Icks A, Lowel H, Meisinger C, et al. High prevalence of undiagnosed diabetes mellitus in Southern Germany: target populations for efficient screening. The KORA survey 2000. Diabetologia. 2003;46(2):182–9.
Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15(7):539–53.
Peters A, Doring A, Ladwig KH, Meisinger C, Linkohr B, et al. Multimorbidity and successful aging: the population-based KORA-Age study. Z Gerontol Geriatr. 2011;44(Suppl 2):41–54.
Petrera A, von Toerne C, Behler J, Huth C, Thorand B, et al. Multiplatform Approach for plasma proteomics: complementarity of Olink Proximity Extension Assay Technology to Mass Spectrometry-based protein profiling. J Proteome Res. 2021;20(1):751–62.
Tunstall-Pedoe H, Kuulasmaa K, Amouyel P, Arveiler D, Rajakangas AM, et al. Myocardial infarction and coronary deaths in the World Health Organization MONICA Project. Registration procedures, event rates, and case-fatality rates in 38 populations from 21 countries in four continents. Circulation. 1994;90(1):583–612.
Alpert JS, Thygesen K, Antman E, Bassand JP. Myocardial infarction redefined–a consensus document of the Joint European Society of Cardiology/American College of Cardiology Committee for the redefinition of myocardial infarction. J Am Coll Cardiol. 2000;36(3):959–69.
Lowel H, Lewis M, Hormann A, Keil U. Case finding, data quality aspects and comparability of myocardial infarction registers: results of a south German register study. J Clin Epidemiol. 1991;44(3):249–60.
Meisinger C, Thorand B, Schneider A, Stieber J, Doring A, et al. Sex differences in risk factors for incident type 2 diabetes mellitus: the MONICA Augsburg cohort study. Arch Intern Med. 2002;162(1):82–9.
World Health Organization, Part III, population survey: section 1, population survey data component: procedures for responders—blood pressure measurement. WHO MONICA Project: MONICA Manual. World Health Organization, Geneva, Switzerland; 1990. p.12–14.
Sun BB, Chiou J, Traylor M, Benner C, Hsu YH, et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature. 2023;622(7982):329–38.
Mbatchou J, Barnard L, Backman J, Marcketta A, Kosmicki JA, et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat Genet. 2021;53(7):1097–103.
Teumer A. Common methods for performing mendelian randomization. Front Cardiovasc Med. 2018;5:51.
Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25.
Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2016;44(D1):D481–487.
Klau S, Jurinovic V, Hornung R, Herold T, Boulesteix AL. Priority-Lasso: a simple hierarchical approach to the prediction of clinical outcome using multi-omics data. BMC Bioinformatics. 2018;19(1):322.
Harrell FE, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982;247(18):2543–6.
Pencina MJ, D’Agostino RB, Sr., Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30(1):11–21.
Pencina MJ, D’Agostino RB, Vasan RS. Statistical methods for assessment of added usefulness of new biomarkers. Clin Chem Lab Med. 2010;48(12):1703–11.
Kolossvary M, deFilippi C, Lu MT, Zanni MV, Fulda ES, et al. Proteomic signature of subclinical coronary artery disease in people with HIV: analysis of the REPRIEVE mechanistic substudy. J Infect Dis. 2022;226(10):1809–22.
Lee CH, Wu MZ, Lui D, Fong C, Ren QW, et al. Prospective associations of circulating thrombospondin-2 level with heart failure hospitalization, left ventricular remodeling and diastolic function in type 2 diabetes. Cardiovasc Diabetol. 2022;21(1):231.
Wallentin L, Eriksson N, Olszowka M, Grammer TB, Hagström E, et al. Plasma proteins associated with cardiovascular death in patients with chronic coronary heart disease: a retrospective study. PLoS Med. 2021;18(1):e1003513.
Herder C, Kannenberg JM, Carstensen-Kirberg M, Strom A, Bonhof GJ, et al. A systemic inflammatory signature reflecting Cross Talk between Innate and adaptive immunity is Associated With Incident Polyneuropathy: KORA F4/FF4 study. Diabetes. 2018;67(11):2434–42.
Lu X, Wang Z, Ye D, Feng Y, Liu M, et al. The role of CXC chemokines in Cardiovascular diseases. Front Pharmacol. 2021;12:765768.
Wei J. Commentary: chemokines in prediabetes and type 2 diabetes: a meta-analysis. Front Immunol. 2021;12:729702.
Zhang D, Contu R, Latronico MV, Zhang J, Rizzi R, et al. MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. J Clin Invest. 2010;120(8):2805–16.
Pan J, Nilsson J, Engström G, De Marinis Y. Elevated circulating follistatin associates with increased risk of mortality and cardiometabolic disorders. Nutr Metabolism Cardiovasc Dis. 2023.
Goncalves I, Hultman K, Duner P, Edsfeldt A, Hedblad B, et al. High levels of cathepsin D and cystatin B are associated with increased risk of coronary events. Open Heart. 2016;3(1):e000353.
Gallo S, Sala V, Gatti S, Crepaldi T. Cellular and molecular mechanisms of HGF/Met in the cardiovascular system. Clin Sci (Lond). 2015;129(12):1173–93.
Bielinski SJ, Berardi C, Decker PA, Larson NB, Bell EJ, et al. Hepatocyte growth factor demonstrates racial heterogeneity as a biomarker for coronary heart disease. Heart. 2017;103(15):1185–93.
Pai JK, Pischon T, Ma J, Manson JE, Hankinson SE, et al. Inflammatory markers and the risk of coronary heart disease in men and women. N Engl J Med. 2004;351(25):2599–610.
Cassidy A, Chiuve SE, Manson JE, Rexrode KM, Girman CJ, et al. Potential role for plasma placental growth factor in predicting coronary heart disease risk in women. Arterioscler Thromb Vasc Biol. 2009;29(1):134–9.
Ligthart S, Sedaghat S, Ikram MA, Hofman A, Franco OH, et al. EN-RAGE: a novel inflammatory marker for incident coronary heart disease. Arterioscler Thromb Vasc Biol. 2014;34(12):2695–9.
Durda P, Sabourin J, Lange EM, Nalls MA, Mychaleckyj JC, et al. Plasma levels of Soluble Interleukin-2 receptor alpha: associations with Clinical Cardiovascular events and genome-wide Association scan. Arterioscler Thromb Vasc Biol. 2015;35(10):2246–53.
Casselbrant A, Fedorowski A, Frantz S, Engstrom G, Wollmer P, et al. Common physiologic and proteomic biomarkers in pulmonary and coronary artery disease. PLoS ONE. 2022;17(3):e0264376.
Matsumoto S, Kishida K, Shimomura I, Maeda N, Nagaretani H, et al. Increased plasma HB-EGF associated with obesity and coronary artery disease. Biochem Biophys Res Commun. 2002;292(3):781–6.
Jin W, Zhao Y, Yan W, Cao L, Zhang W, et al. Elevated circulating interleukin-27 in patients with coronary artery disease is associated with dendritic cells, oxidized low-density lipoprotein, and severity of coronary artery stenosis. Mediators Inflamm. 2012;2012:506283.
Oh TJ, Ahn CH, Kim BR, Kim KM, Moon JH, et al. Circulating sortilin level as a potential biomarker for coronary atherosclerosis and diabetes mellitus. Cardiovasc Diabetol. 2017;16(1):92.
Hwang JJ, Yang WS, Chiang FT, Chen MF, Lin HJ, et al. Association of circulating matrix metalloproteinase-1, but not adiponectin, with advanced coronary artery disease. Atherosclerosis. 2009;204(1):293–7.
Carvalho VMF, Oliveira PSS, Albuquerque APB, Rego M, Rosa MMD, et al. Decreased serum levels of Soluble Oncostatin M receptor (sOSMR) and glycoprotein 130 (sgp130) in patients with coronary artery disease. Arq Bras Cardiol. 2023;120(4):e20220326.
Demyanets S, Speidl WS, Tentzeris I, Jarai R, Katsaros KM, et al. Soluble ST2 and interleukin-33 levels in coronary artery disease: relation to disease activity and adverse outcome. PLoS ONE. 2014;9(4):e95055.
Lira-Junior R, Bostrom EA, Gustafsson a on behalf of the PAROKRANK steering committee. Periodontitis is associated to increased systemic inflammation in postmyocardial infarction patients. Open Heart. 2021;8(2):e001674.
Ndongo-Thiam N, de Sallmard G, Kastrup J, Miossec P. Levels of soluble osteoclast-associated receptor (sOSCAR) in rheumatoid arthritis: link to disease severity and cardiovascular risk. Ann Rheum Dis. 2014;73(6):1276–7.
Kong P, Cui ZY, Huang XF, Zhang DD, Guo RJ, et al. Inflammation and atherosclerosis: signaling pathways and therapeutic intervention. Signal Transduct Target Ther. 2022;7(1):131.
Hansson GK, Libby P, Schonbeck U, Yan ZQ. Innate and adaptive immunity in the pathogenesis of atherosclerosis. Circ Res. 2002;91(4):281–91.
McCarthy CP, van Kimmenade RRJ, Gaggin HK, Simon ML, Ibrahim NE, et al. Usefulness of multiple biomarkers for Predicting Incident Major adverse cardiac events in patients who underwent Diagnostic Coronary Angiography (from the Catheter sampled blood archive in Cardiovascular diseases [CASABLANCA] Study). Am J Cardiol. 2017;120(1):25–32.
Acknowledgements
We thank all participants for their long-term commitment to the KORA study, the staff for data collection and research data management and the members of the KORA Study Group (https://www.helmholtz-munich.de/en/epi/cohort/kora) who are responsible for the design and conduct of the KORA studies.
Funding
Open Access funding enabled and organized by Projekt DEAL. HL was supported by a scholarship under the State Scholarship Fund by the China Scholarship Council (File No. 201906380066). The KORA study was initiated and financed by the Helmholtz Zentrum München – German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. Data collection in the KORA study is done in cooperation with the University Hospital of Augsburg. The present study was supported with proteomics data by Helmholtz Institute for Metabolic, Obesity and Vascular Research – Project Initiative 2018 (HI-MAG). The German Diabetes Center is funded by the German Federal Ministry of Health (Berlin, Germany) and the Ministry of Culture and Science of the state North Rhine-Westphalia (Düsseldorf, Germany) and receives additional funding from the German Federal Ministry of Education and Research (BMBF) through the German Center for Diabetes Research (DZD e.V.).
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
HL and BT conceptualised the research question. HL drafted the analysis plan, performed the statistical analysis and wrote the manuscript. BT and MTH contributed to revising the manuscript and interpreting the results. AH provided statistical analysis advice. A. Petrera, SMH, WR, CH, WK, A. Peters and BT contributed data. All authors were involved in the review and final approval of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was performed in accordance with the Declaration of Helsinki. All study methods were approved by the Ethics Committee of the Bavarian Chamber of Physicians, Munich (KORA S4: EC No. 99186) and the Bavarian Medical Association (KORA-Age: EC No. 08094). Written informed consent was obtained from all study participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Luo, H., Huemer, MT., Petrera, A. et al. Association of plasma proteomics with incident coronary heart disease in individuals with and without type 2 diabetes: results from the population-based KORA study. Cardiovasc Diabetol 23, 53 (2024). https://doi.org/10.1186/s12933-024-02143-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12933-024-02143-z