Abstract
Background
The high heterogeneity in the symptoms and severity of COVID-19 makes it challenging to identify high-risk patients early in the disease. Cardiometabolic comorbidities have shown strong associations with COVID-19 severity in epidemiologic studies. Cardiometabolic protein biomarkers, therefore, may provide predictive insight regarding which patients are most susceptible to severe illness from COVID-19.
Methods
In plasma samples collected from 343 patients hospitalized with COVID-19 during the first wave of the pandemic, we measured 92 circulating protein biomarkers previously implicated in cardiometabolic disease. We performed proteomic analysis and developed predictive models for severe outcomes. We then used these models to predict the outcomes of out-of-sample patients hospitalized with COVID-19 later in the surge (N = 194).
Results
We identified a set of seven protein biomarkers predictive of admission to the intensive care unit and/or death (ICU/death) within 28 days of presentation to care. Two of the biomarkers, ADAMTS13 and VEGFD, were associated with a lower risk of ICU/death. The remaining biomarkers, ACE2, IL-1RA, IL6, KIM1, and CTSL1, were associated with higher risk. When used to predict the outcomes of the future, out-of-sample patients, the predictive models built with these protein biomarkers outperformed all models built from standard clinical data, including known COVID-19 risk factors.
Conclusions
These findings suggest that proteomic profiling can inform the early clinical impression of a patient’s likelihood of developing severe COVID-19 outcomes and, ultimately, accelerate the recognition and treatment of high-risk patients.
Similar content being viewed by others
Background
The COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in millions of deaths and persists as a global health threat [1]. COVID-19 causes a spectrum of symptoms, from mild upper respiratory tract infection to severe acute respiratory distress syndrome (ARDS) and death [2,3,4]. Most patients who develop severe COVID-19 deteriorate after one week of symptom onset [5]. As existing viral modifying therapies, such as monoclonal antibodies, are most effective when given before patients develop critical symptoms [6], early recognition is crucial to the triage and treatment of severe cases.
Previous studies of patients hospitalized with COVID-19 have consistently found significant associations between comorbidities and poor COVID-19 outcomes [7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29]. A cross-sectional study of over half a million hospitalized adults with COVID-19 found that 95% had at least one underlying medical condition and observed a strong dose–response association between the total number of underlying conditions (e.g., hypertension, obesity, lipid disorder, diabetes, and coronary artery disease) and risk of severe COVID-19 illness. Compared to patients with no documented underlying medical conditions, having one medical condition (7% of patients) was associated with 1.5 times higher risk of death, and having more than 10 conditions (17% of patients) was associated with a fourfold increase in the risk of death [7]. While little progress has been made in the development of tests that reliably predict which patients will suffer poor outcomes from COVID-19 (e.g., need for intensive care, assisted ventilation, septic shock, multiorgan system dysfunction, or death) [10], the presence of certain comorbid conditions, including higher body mass index (BMI) [7, 11,12,13,14,15,16], coronary artery disease [17,18,19], chronic kidney disease [20,21,22,23,24,25], and type 2 diabetes [7, 8, 21, 26,27,28,29], can help identify the patients most susceptible to severe forms of COVID-19.
Circulating protein biomarkers reflect underlying physiologic or disease states and can serve as measurable indicators of disease severity for prognostication in clinical practice. Interrogating the relationship between COVID-19 outcomes and protein biomarkers previously implicated in cardiometabolic disease may yield actionable insights regarding how common cardiometabolic comorbidities contribute to COVID-19 pathogenesis and, ultimately, improve our ability to predict which patients are most likely to suffer poor outcomes.
Methods
Study design
During the first wave of the pandemic (March 10 to June 1, 2020), we collected discarded blood samples from patients who presented to care (i.e., first contact with the healthcare system) with COVID-19 symptoms and were subsequently hospitalized with PCR-confirmed SARS-CoV-2 infection at Massachusetts General Hospital (MGH), a large academic hospital in Boston, Massachusetts. In these samples, we measured 92 protein biomarkers in the Olink Target 96 Cardiovascular II panel [30]. We performed proteomic analysis and developed predictive models for the combined outcome of death or admission to the intensive care unit (ICU/death) within 28 days of presentation to care in patients hospitalized early in the surge, between March 10 and April 21, 2020 (“in-sample” group; n = 343). We then used these models to predict the outcomes in a separate sample of patients hospitalized with COVID-19 later in the surge, between April 22 and June 1, 2020 (“out-of-sample” group; n = 194).
Data collection
Details of the hospitalization, past medical history, and demographic characteristics were collected for each patient by manual chart reviews. Hospital laboratory tests performed within three days of presentation to care were retrieved electronically through the Enterprise Data Warehouse, a repository derived from electronic health records. We excluded hospital laboratory tests with a missing rate greater than 30% and imputed those remaining with the median of the non-missing values (mean missing rate = 13%). For each patient, we generated the CURB-65 score, a risk score for pneumonia severity [31]. The CURB-65 score ranges from 0 to 5 based on the presence of the following factors: (1) confusion (defined as Glasgow Coma Scale < 15), (2) blood urea nitrogen > 19 mg/dL, (3) respiratory rate ≥ 30, (4) systolic blood pressure < 90 mmHg or diastolic blood pressure ≤ 60 mmHg, and (5) age ≥ 65. Study procedures were approved by the Mass General Brigham (formerly Partners) Human Research Committee, the governing institutional review board at MGH.
Plasma collection and proteomic assays
Blood samples were collected in EDTA tubes and processed within three days of collection in a biosafety level 2 + laboratory. Whole blood was centrifuged at 2800 rpm for 10 min. Plasma samples were extracted and stored at −80 °C. Prior to plating, samples were treated for two hours with 1% TritonX-100 at room temperature for virus inactivation. The Olink reagents were based on Proximity Extension Assay technology. Measurements were translated into normalized protein expression (NPX) units. Sample plates passed quality control if the standard deviations of internal controls were < 0.20 NPX and individual samples were < 0.30 NPX from the median value of the internal controls [30]. A total of 537 samples (95%) passed quality control. All 92 protein biomarkers were detected with intra-assay coefficient of variance (CV) < 15% (mean 4%) and inter-assay CV < 30% (mean 10%).
Statistical analysis
We performed all statistical analyses in Python version 3.9.2 and R version 4.0.5. To account for non-normality in the raw data, we applied rank-based inverse normal transformation for feature selection, modeling, and P value calculation. To preserve interpretability, the odds ratios (ORs) shown in the volcano plot and correlation matrix were calculated after standardizing the data to have a mean of 0 and standard deviation of 1. To account for multiple testing, the threshold P < 0.05/116 hospital laboratory tests and protein biomarkers = 4 × 10–4 was used to determine significance.
To uncover mechanistic links, we performed a core enrichment analysis using QIAGEN Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA) by matching a proteomic dataset, consisting of P values and ORs for each protein biomarker, with curated content of the Ingenuity Knowledge Base. We generated reports for molecular networks and canonical pathways, highlighting upstream regulators and pathways relevant to the observed changes, along with their predicted impact on downstream biological or disease processes.
Predictive modeling
To identify the subset of variables with the greatest predictive value for the logistic regression model, we ranked all variables by the frequency with which they were selected by LASSO (least absolute shrinkage and selection operator) regression when repeated across separate, random subsets of the in-sample data and different shrinkage parameters [32]. To identify the best model, we used an all-possible-regressions approach with these top-ranked variables. We used fivefold stratified cross-validation within the in-sample data to test the models formed by every possible combination of top-ranked variables. We then plotted the average area under the receiver operating characteristic curve (AUC), with the models sorted by the number of variables they included. The model that formed the elbow at which the plot of increasing AUC versus model size began to plateau was considered the best model, achieving the greatest performance without over-fitting the in-sample data. We performed this analysis with and without the 92 protein biomarkers and compared the AUCs of the best models built with and without the biomarkers using a two-sided DeLong test [33]. We repeated the modeling process using random forest models and using the Boruta algorithm to initially rank the variables by significance [34]. Data from the out-of-sample patients was not used at any point in the feature selection and modeling process.
Results
Characteristics of the patients
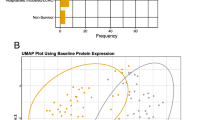
Figure 1 shows the baseline characteristics of patients in the in-sample group and out-of-sample group. Table 1 shows the patient characteristics stratified by the ICU/death outcome in each group. Compared to the in-sample group, the out-of-sample group had a similar proportion of patients who died, but a smaller proportion of patients who were admitted to the ICU (Fig. 1). The out-of-sample group had a larger proportion of self-reported non-Hispanic White and non-Hispanic Black/African-American patients, a smaller proportion of self-reported Hispanic patients, a lower average BMI, higher levels of D-dimer, lower levels of lactate dehydrogenase, and lower levels of creatine kinase relative to the in-sample group. Patients who died or were admitted to the ICU had a higher average BMI, higher average CURB-65 score, and higher levels of D-dimer, LDH, C-reactive protein (CRP), and ferritin compared to those alive and not admitted to the ICU after 28 days of presentation to care (Table 1).
Characteristics of in-sample and out-of-sample patients. Blood samples were collected from 537 patients hospitalized with COVID-19 during an early surge in the outbreak, between March 10th and June 1st of 2020. Data from patients who were hospitalized early in the surge (before April 22, 2020) was used to analyze the cardiometabolic protein biomarkers and develop logistic regression and random forest models for severe outcomes. These patients comprised the in-sample group (shown in grey). These models were then used to predict the outcomes of the out-of-sample patients (shown in gold) who were hospitalized later in the surge (starting April 22, 2020). The in-sample and out-of-sample patients were compared across various demographic and clinical variables using a two-sided t-test for continuous variables and chi-square test for categorical variables. All race/ethnicity categories were self-reported. BMI categorization: < 18.5 kg/m2 for underweight, 18.5–24.9 kg/m2 for normal weight, 25.0–29.9 kg/m2 for overweight, and ≥ 30.0 kg/m2 for obese. SD Standard deviation, AA African American, BMI body mass index, CAD coronary artery disease, COPD chronic obstructive pulmonary disease, CRP C-reactive protein, LDH lactate dehydrogenase
Cardiometabolic protein biomarkers
To evaluate the relative importance of each of the 92 protein biomarkers and the 24 hospital laboratory tests with respect to ICU/death, we built a logistic regression model, adjusted for age, gender, BMI, and self-reported race/ethnicity, for each biomarker and hospital laboratory test in the in-sample group (Additional file 1: Table S1). The protein biomarkers comprised 31 of the 36 significant associations (P < 4 × 10–4) with ICU/death (Fig. 2). The five hospital laboratory tests with significant associations included LDH, CRP, procalcitonin, aspartate aminotransferase, and alanine transaminase, with standardized odds ratios (ORs) ranging from 1.8 to 3.2 (Additional file 1: Fig. S1).
Volcano plot of the 92 cardiometabolic biomarkers and 24 hospital laboratory tests. The plot includes, for each protein biomarker and hospital lab, the odds ratio on the x-axis and P value (-log10) on the y-axis resulting from a logistic regression model with ICU/death as the outcome, adjusted for the covariates: age, gender, BMI, and self-reported race/ethnicity. To account for non-normality, the P values were calculated after applying rank-based inverse normal transformation. To preserve interpretability, the odds ratios were calculated from the data standardized to have a mean of 0 a standard deviation of 1. The threshold P < 0.05/116 hospital laboratory tests and protein biomarkers = 4 × 10–4 was used to identify significant results (shown in red). Nominally significant results (P < 0.05) are shown in green. SD Standard deviation, CRP C-reactive protein, LDH lactate dehydrogenase
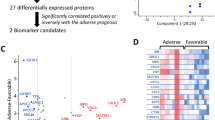
We evaluated how the protein biomarkers that were significantly associated with ICU/death correlated with one another and with the hospital laboratory tests, comorbidities, and demographics (Fig. 3). The largest cluster included 16 biomarkers which were positively associated with ICU/death (with standardized ORs ranging from 1.4 to 5.2) and showed positive correlations with type 2 diabetes, chronic kidney disease, and cardiac disease (Fig. 3: Box A). This cluster also showed significant positive correlations with troponin, blood urea nitrogen, creatinine, procalcitonin, and D-dimer (Fig. 3: Box B) and negative correlations with estimated glomerular filtration rate, albumin, hematocrit, and hemoglobin (Fig. 3: Box C). A smaller cluster of five biomarkers (ADAMTS13, SCF, FABP2, VEGFD, and TGM2) was negatively associated with ICU/death and negatively correlated with the majority of the hospital laboratory tests (Fig. 3: Box D). Among all 36 significant biomarkers and hospital laboratory tests, this cluster included the only markers associated with lower risk of ICU/death (with standardized ORs ranging from 0.40 to 0.64). Canonical pathways identified by IPA included the tumor microenvironment, IL-10 signaling, airway pathology, granulocyte adhesion, and wound healing. Implicated functional networks included cardiovascular and organismal development, lipid metabolism, and protein synthesis (Additional file 1: Tables S2 and S3).
Hierarchical clustering and correlation matrix with significant cardiometabolic biomarkers. A heatmap (top left) and correlation matrix (top right and bottom) for the 31 protein biomarkers significantly associated with ICU/death (P < 0.05/116 hospital laboratory tests and biomarkers = 4 × 10–4). The correlation matrix shows how the protein biomarkers, ordered based on hierarchical clustering, correlate with one another (top right) and how they correlate with the demographic factors, clinical variables, and hospital laboratory tests (bottom). The color reflects the magnitude and direction of the Pearson correlation coefficient. The cells corresponding to correlations with P > 0.05 were left blank. The P values and odds ratios (OR) reported for the association of each variable with ICU/death are the same as those shown in Fig. 2. Box A shows the association of the largest cluster, comprised of 16 biomarkers, with type 2 diabetes, chronic kidney disease (CKD), and cardiac disease. Boxes B and C show how this cluster correlates with the hospital labs. Finally, Box D shows correlations between the hospital laboratory tests and a smaller cluster, comprising the five biomarkers that were negatively associated with ICU/death. SD Standard deviation, CI confidence interval, AA African American, COPD chronic obstructive pulmonary disease, CAD coronary artery disease, HFpEF heart failure with preserved ejection fraction, HFrEF heart failure with reduced ejection fraction, BUN blood urea nitrogen, ERS erythrocyte sedimentation rate, LDH lactate dehydrogenase, AST aspartate aminotransferase, WBC white blood cells, CRP C-reactive protein, ALC absolute lymphocyte count, eGFR estimated glomerular filtration rate
Prediction in out-of-sample patients
For both the logistic regression and random forest, the models built with the protein biomarkers outperformed the models built without the biomarkers in the out-of-sample patients (Fig. 4). The best-performing models consisted of a common set of nine variables (Table S4), which included two hospital laboratory measurements, procalcitonin and LDH (both of which were included in the best models without the biomarkers), and seven biomarkers: IL-1RA, CTSL1, ADAMTS13, VEGFD, KIM1, ACE2, and IL6 (Fig. 4A). The AUCs of the best models built with these nine variables were greater than that of the best models built without the protein biomarkers (logistic regression: 0.82 versus 0.70; P = 0.001; random forest: 0.83 versus 0.69; P = 3 × 10–5). We continued to observe superior performance by the models built with the protein biomarkers when excluding patients with more than 14 days between presentation to care and sample collection date (Additional file 1: Fig. S2 and Table S5), when excluding patients with sample collection on or after the ICU admission date (Additional file 1: Fig. S3), when randomly splitting patients into the in-sample and out-of-sample group (Additional file 1: Fig. S4), when stratifying patients by CURB-65 score and adjusting the models by CURB-65 score (Additional file 1: Table S6), and when directly adjusting each model by variables such as age, sex, and relevant comorbidities (e.g., BMI, COPD, asthma, type 2 diabetes, CKD, cardiac disease, smoking history, cancer history). Despite the loss of power, we see a similar difference in AUC between the model without biomarkers and the model with biomarkers (0.70 versus 0.79, p = 0.06) when excluding patients with a sample collection date greater than three days following presentation to care. The models with the protein biomarkers also outperformed the models without biomarkers in age-stratified and gender-stratified analyses (Additional file 1: Fig. S5 and S6). When evaluating the model performance in the two most prevalent self-reported categories of race/ethnicity, the improvement in performance of the model with biomarkers was greater in the Hispanic population than the non-Hispanic White population (Additional file 1: Fig. S7). We repeated the analysis using ICU admission as the outcome and death as the outcome. The results for the ICU admission outcome were similar to those for the combined ICU/death outcome (Fig. S8 and Table S7), while the results for the death outcome showed no significant difference in performance between the models built with and without the biomarkers (Additional file 1: Fig. S9 and Table S8).
Prediction of ICU/death outcome in out-of-sample patients. A Violin plots for the set of seven cardiometabolic protein biomarkers that were included in the best model with biomarkers for both logistic regression and random forest. The figure depicts the distribution and box plot of these seven biomarkers, stratified by the ICU/death outcome, in the in-sample patient population. The P values shown for each biomarker are based on the rank-inverse normalized data, while the odds ratios (OR) are based on the data standardized to have a mean of 0 and standard deviation of 1. B The predictive performance of the best models with and without biomarkers in the out-of-sample patients. The figure shows the receiver operating characteristic curve and corresponding area under the curve (AUC) for the best logistic regression (left) and random forest (right) models with biomarkers (gold) and without biomarkers (bronze) in the out-of-sample patients. The best model with biomarkers, for both the logistic regression and random forest, included the same set of seven biomarker, shown in (A), along with two hospital labs: procalcitonin and LDH. All models were developed and trained using only the in-sample data. Thrombotic thrombocytopenic purpura, TTP; acute respiratory distress syndrome, ARDS
Discussion
We identified protein biomarkers previously implicated in cardiometabolic disease that were significantly associated with severe illness from COVID-19, shedding light on biological pathways involved in COVID-19 pathology. We demonstrated that these protein biomarkers, measured early in the disease course, were more predictive of ICU admissions or death than established clinical risk factors. These findings suggest that proteomic profiling could improve the triage and treatment of patients hospitalized with COVID-19.
We found a set of seven protein biomarkers (IL6, IL-1RA, KIM1, ACE2, CTSL1, ADAMTS13, and VEGFD), along with two hospital laboratory tests (procalcitonin and LDH), that were predictive of ICU/death. These circulating biomarkers are likely related to host and viral factors influencing disease, including inflammation (IL6, IL-1RA) [35, 36], thrombosis (ADAMTS13, VEGFD) [37, 38], and viral entry (KIM1, ACE2, CTSL1) [39,40,41]. Elevated inflammatory markers, including IL-6 [35, 42,43,44], CRP, ferritin, and D-dimer have been reported in severe COVID-19 [45]. Dexamethasone, an anti-inflammatory medication, and IL-6 receptor antagonists are current COVID-19 therapies shown to reduce the risk of poor outcomes in critically ill patients [46, 47].
LDH, D-dimer, fibrinogen, CRP, and low platelets, markers of thrombotic risk, have been reported to be associated with poor prognosis in COVID-19 [48]. This observation is in keeping with the association between lower ADAMTS13, an enzyme that degrades von Willebrand factor, and poor outcomes found in our study and other reports [37]. Low levels of ADAMTS13 have also been described in thrombotic thrombocytopenic purpura and syndromes of thrombotic microangiopathy caused by infection [49]. Microangiopathic thrombosis has been seen in autopsies of patients who have died of COVID-19, similar to what has been observed in other ARDS-causing diseases [50].
Three of the identified biomarkers, KIM1, ACE2, and CTSL, are involved in host-virus interactions. KIM1, an indicator of renal insults, plays a role in viral entry and regulation of the host immune response to viral infections [51]. ACE2, the cellular receptor for SARS-CoV-2 [40, 42], undergoes shedding, leading to circulating ACE2, a biomarker of cardiovascular disease, diabetes, and death in patients with and without COVID-19 [52, 53]. The association of ACE2 with severity is supported by a recently reported rare genetic variant that is associated with a 37% reduction in ACE2 expression and a 40% reduction in risk of severe COVID-19 [54]. Finally, CTSL is one of the lysosomal proteases that can cleave the SARS-CoV-2 spike protein, a step necessary for cellular entry [41, 55].
Previous studies have reported hospital laboratory tests and clinical characteristics associated with severe COVID-19 [5, 35, 36, 44, 45, 56,57,58,59,60,61,62]. These studies used both prospective and retrospective analyses of clinical variables, imaging findings, and laboratory values predictive of severity. Laboratory values such as IL6 [35, 44], IL-1RA, IL10 [36], D-dimer [5, 45, 61], and troponin [5, 60, 62] were associated with disease severity. Clinical characteristics, such as BMI, age [5, 63], history of renal failure, cardiovascular and cerebrovascular disease [60, 63], were also associated with worse outcomes. Our study shows the best predictive model included many of these previously reported risk factors when the protein biomarkers were not included (Table S4). However, when the protein biomarkers were included in the model-building process, these risk factors were replaced with the set of seven biomarkers, resulting in models that significantly outperformed all models developed from the clinical features and laboratory tests alone, suggesting that the biomarkers provide unique predictive value not captured by patient data and hospital lab values. The protein biomarkers replaced known clinical risk factors for severe illness that had been selected in the model built without biomarkers (i.e., BMI, D-dimer, CRP, ALC, and troponin). Notably, BMI was replaced by IL-1RA, a biomarker that was strongly correlated with BMI (Fig. 3). IL-1RA, known to be highly expressed in white adipose tissue [64] and upregulated during inflammation, could serve as a better proxy than BMI for obesity-driven COVID-19 risk. D-dimer was another previously reported COVID-19 risk factor [5, 45, 61] that was selected in the model without protein biomarkers but replaced in the model with biomarkers. D-dimer was positively correlated with IL6, IL-1RA, KIM1, ACE2, and CTSL1 (Fig. 3: Box B) and negatively correlated with ADAMTS13 and VEGFD (Fig. 3: Box D). The replacement of D-dimer in the final model with these seven protein biomarkers suggests that the predictive value provided by D-dimer was captured by this combination of biomarkers. Nevertheless, the utility of these protein biomarkers for the purpose of risk stratification in real-world clinical settings will need to be prospectively assessed in the context of their practical considerations (e.g., cost, speed of results reporting, and access to testing) for them to be appropriately incorporated in the clinical evaluation of patients hospitalized with COVID-19.
We recognize that standards of care and resource availability evolved quickly during the first wave of the pandemic. As data on the efficacy and side effects of COVID-19 therapies accrued, the use of remdesivir and dexamethasone increased, while the use of hydroxychloroquine decreased. MGH hospital guidelines did not recommend routine systemic anticoagulation for patients with COVID-19 during the pandemic and did not recommend the use of steroids, such as dexamethasone, until the publication of the RECOVERY study [65], after the recruitment period of this study. Prone positioning, applied heterogeneously early in the pandemic, eventually became standard of care. It is possible that these exogenous factors contributed to differences in outcomes between the in-sample and out-of-sample cohorts. We expect that, as the SARS-CoV-2 virus mutates, the virulence pathways and host responses may change, as noted by both the delta and omicron variants [66]. The patients hospitalized with COVID-19 today are generally younger and consist of both unvaccinated and vaccinated patients with breakthrough infections or repeat infections. Emerging COVID-19 therapies and medication exposures for preexisting medical conditions could influence the proteomic profile of patients and its association with COVID-19 severity.
By evaluating the models in a sample separate from that used to develop the models, we showed that the predictive value of the biomarkers was robust to changes in clinical protocols and the patient characteristics during the highly dynamic study period. Compared to studies that develop and test predictive models within the same patient population, our approach provided a more rigorous assessment of the generalizability of our models and the conclusions derived from our analysis. As one of the largest proteomic analyses performed in COVID-19 patients, we were able to conduct age-stratified, gender-stratified, and race/ethnicity-stratified analyses, demonstrating the strong performance of the model with the protein biomarkers across various demographic strata (Additional file 1: Fig. S5, S6, and S7).
Similar to previous COVID-19 analyses [67,68,69], this study was limited by the precision with which COVID-19 severity could be captured and COVID-19 related outcomes could be tracked. We used ICU admission and death as proxies for severe illness from COVID-19; however, patients may have died or been admitted to the ICU for reasons independent of their COVID-19 status. Further, patients who died after the 28-day follow-up period or outside the hospital or at other hospitals would not have been captured in our study. This underestimate of the true case-count for the death outcome may have biased results towards the null (Additional file 1: Fig. S9). Another limitation was that the hospital laboratory tests and protein biomarkers were not measured at the same time for all patients and the time between symptom onset and blood sample collection was not uniform across all patients. Despite this, we observed similar results when excluding patients with sample collection dates that were on or after the date of ICU admission or greater than three days or 14 days following presentation to care (Additional file 1: Fig. S2, Fig. S3, and Table S5). Finally, by only collecting discarded blood samples at a single time point, we were unable to perform longitudinal analyses; however, biomarkers that can be interpreted with single timepoint measurements may be more useful in clinical settings where only one lab draw is available. Our retrospective proteomic analysis shows that protein biomarkers improve prediction of severe outcomes over clinical biomarkers and risk factors that are routinely measured or obtained from hospitalized patients with COVID-19.
Conclusion
In this study, we identified a set of protein biomarkers that yield both mechanistic insight regarding how cardiometabolic disease contributes to COVID-19 pathology, as well as predictive value regarding which patients have the highest risk for severe outcomes. If considered early in the clinical evaluation of patients with COVID-19, these insights can help clinicians estimate a patient’s cardiometabolic-driven risk, which, in turn, can inform downstream decisions regarding how to stratify patients across pathways of clinical care (e.g., in-hospital observation or early admission to ICU) and whether to institute treatments that reduce the risk of poor outcomes, such as monoclonal antibodies or novel antiviral therapies.
Availability of data and materials
The datasets generated and analyzed during the current study are not publicly available but may be made available upon reasonable request.
Abbreviations
- AA:
-
African American
- ACE2:
-
Angiotensin converting enzyme 2
- ADAMTS13:
-
A disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13
- ALC:
-
Absolute lymphocyte count
- ARDS:
-
Acute respiratory distress syndrome
- AST:
-
Aspartate aminotransferase
- AUC:
-
Area under the curve
- BMI:
-
Body mass index
- BUN:
-
Blood urea nitrogen
- CAD:
-
Coronary artery disease
- CKD:
-
Chronic kidney disease
- COPD:
-
Chronic obstructive pulmonary disease
- COVID-19:
-
Coronavirus disease 19
- CRP:
-
C-reactive protein
- CTSL1:
-
Cathepsin L1
- CV:
-
Coefficient of variance
- EGFR:
-
Estimated glomerular filtration rate
- ERS:
-
Eerythrocyte sedimentation rate
- HFpEF:
-
Heart failure with preserved ejection fraction
- HFrEF:
-
Heart failure with reduced ejection fraction
- ICU:
-
Intensive care unit
- IL-1RA:
-
Interleukin 1 receptor antagonist
- IL6:
-
Interleukin 6
- IPA:
-
Ingenuity Pathway Analysis
- KIM1:
-
Kidney injury molecule 1
- LASSO:
-
Least absolute shrinkage and selection operator
- LDH:
-
Lactate dehydrogenase
- MGH:
-
Massachusetts General Hospital
- NPX:
-
Normalized protein expression
- OR:
-
Odds ratio
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- SD:
-
Standard deviation
- TTP:
-
Thrombotic thrombocytopenic purpura
- VEGFD:
-
Vascular endothelial growth factor D
- WBC:
-
White blood cells
References
Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–4.
Cai Q, Chen F, Wang T, Luo F, Liu X, Wu Q, He Q, Wang Z, Liu Y, Liu L, et al. Obesity and COVID-19 severity in a designated hospital in Shenzhen, China. Diabetes Care. 2020. https://doi.org/10.2337/dc20-0576.
Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, Zimmer T, Thiel V, Janke C, Guggemos W, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382(10):970–1.
Bai Y, Yao L, Wei T, Tian F, Jin DY, Chen L, Wang M. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020. https://doi.org/10.1001/jama.2020.2565.
Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, Ma K, Xu D, Yu H, Wang H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368: m1091.
Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med. 2020;384(3):238–51.
Kompaniyets L, Pennington AF, Goodman AB, Rosenblum HG, Belay B, Ko JY, Chevinsky JR, Schieber LZ, Summers AD, Lavery AM, et al. underlying medical conditions and severe illness among 540,667 adults hospitalized with Covid-19, March 2020-March 2021. Prev Chronic Dis. 2021;18:E66.
Harrison SL, Fazio-Eynullayeva E, Lane DA, Underhill P, Lip GYH. Comorbidities associated with mortality in 31,461 adults with COVID-19 in the United States: a federated electronic medical record analysis. PLoS Med. 2020;17(9): e1003321.
De Giorgi A, Fabbian F, Greco S, Di Simone E, De Giorgio R, Passaro A, Zuliani G, Manfredini R. Prediction of in-hospital mortality of patients with SARS-CoV-2 infection by comorbidity indexes: an Italian internal medicine single center study. Eur Rev Med Pharmacol Sci. 2020;24(19):10258–66.
Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323(13):1239–42.
Leong A, Cole JB, Brenner LN, Meigs JB, Florez JC, Mercader JM. Cardiometabolic risk factors for COVID-19 susceptibility and severity: a Mendelian randomization analysis. PLoS Med. 2021;18(3): e1003553.
Lighter J, Phillips M, Hochman S, Sterling S, Johnson D, Francois F, Stachel A. Obesity in patients younger than 60 years is a risk factor for COVID-19 hospital admission. Clin Infect Dis. 2020;71:896–7.
Hur K, Price CPE, Gray EL, Gulati RK, Maksimoski M, Racette SD, Schneider AL, Khanwalkar AR. Factors associated with intubation and prolonged intubation in hospitalized patients with COVID-19. Otolaryngol Head Neck Surg. 2020;163(1):170–8.
Simonnet A, Chetboun M, Poissy J, Raverdy V, Noulette J, Duhamel A, Labreuche J, Mathieu D, Pattou F, Jourdain M. High prevalence of obesity in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) requiring invasive mechanical ventilation. Obesity (Silver Spring). 2020;28(7):1195–9.
Kalligeros M, Shehadeh F, Mylona EK, Benitez G, Beckwith CG, Chan PA, Mylonakis E. Association of obesity with disease severity among patients with coronavirus disease 2019. Obesity (Silver Spring). 2020;28(7):1200–4.
Palaiodimos L, Kokkinidis DG, Li W, Karamanis D, Ognibene J, Arora S, Southern WN, Mantzoros CS. Severe obesity, increasing age and male sex are independently associated with worse in-hospital outcomes, and higher in-hospital mortality, in a cohort of patients with COVID-19 in the Bronx New York. Metabolism. 2020;108: 154262.
Chen R, Liang W, Jiang M, Guan W, Zhan C, Wang T, Tang C, Sang L, Liu J, Ni Z, et al. Risk factors of fatal outcome in hospitalized subjects with coronavirus disease 2019 from a nationwide analysis in China. Chest. 2020;158(1):97–105.
Yang J, Zheng Y, Gou X, Pu K, Chen Z, Guo Q, Ji R, Wang H, Wang Y, Zhou Y. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int J Infect Dis. 2020;94:91–5.
Zheng Z, Peng F, Xu B, Zhao J, Liu H, Peng J, Li Q, Jiang C, Zhou Y, Liu S, et al. Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect. 2020;81(2):e16–25.
Garg S, Kim L, Whitaker M, O’Halloran A, Cummings C, Holstein R, Prill M, Chai SJ, Kirley PD, Alden NB, et al. Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019—COVID-NET, 14 States, March 1–30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(15):458–64.
Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, Barnaby DP, Becker LB, Chelico JD, Cohen SL, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020;323(20):2052–9.
Akalin E, Azzi Y, Bartash R, Seethamraju H, Parides M, Hemmige V, Ross M, Forest S, Goldstein YD, Ajaimy M, et al. Covid-19 and kidney transplantation. N Engl J Med. 2020;382:2475–7.
Gold JAW, Wong KK, Szablewski CM, Patel PR, Rossow J, da Silva J, Natarajan P, Morris SB, Fanfair RN, Rogers-Brown J, et al. Characteristics and clinical outcomes of adult patients hospitalized with COVID-19—Georgia, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69(18):545–50.
Hirsch JS, Ng JH, Ross DW, Sharma P, Shah HH, Barnett RL, Hazzan AD, Fishbane S, Jhaveri KD. Acute kidney injury in patients hospitalized with COVID-19. Kidney Int. 2020;98(1):209–18.
Myers LC, Parodi SM, Escobar GJ, Liu VX. Characteristics of hospitalized adults with COVID-19 in an integrated health care system in California. JAMA. 2020;323(21):2195–8.
Zhu L, She ZG, Cheng X, Qin JJ, Zhang XJ, Cai J, Lei F, Wang H, Xie J, Wang W, et al. Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing type 2 diabetes. Cell Metab. 2020;31(6):1068-1077.e1063.
Bode B, Garrett V, Messler J, McFarland R, Crowe J, Booth R, Klonoff DC. Glycemic characteristics and clinical outcomes of COVID-19 patients hospitalized in the United States. J Diabetes Sci Technol. 2020;14(4):813–21.
Chen Y, Yang D, Cheng B, Chen J, Peng A, Yang C, Liu C, Xiong M, Deng A, Zhang Y, et al. Clinical characteristics and outcomes of patients with diabetes and COVID-19 in association with glucose-lowering medication. Diabetes Care. 2020;43(7):1399–407.
Fadini GP, Morieri ML, Longato E, Avogaro A. Prevalence and impact of diabetes among people infected with SARS-CoV-2. J Endocrinol Invest. 2020;43:867–9.
Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, Eriksson A, Rennel Dickens E, Ohlsson S, Edfeldt G, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE. 2014;9(4): e95192.
Guo J, Zhou B, Zhu M, Yuan Y, Wang Q, Zhou H, Wang X, Lv T, Li S, Liu P, et al. CURB-65 may serve as a useful prognostic marker in COVID-19 patients within Wuhan, China: a retrospective cohort study. Epidemiol Infect. 2020;148: e241.
Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc (Series B). 1996;58:267–88.
DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–45.
Kursa MB, Jankowski A, Rudnicki WR. Boruta–a system for feature selection. Fund Inform. 2010;101(4):271–85.
Galván-Román JM, Rodríguez-García SC, Roy-Vallejo E, Marcos-Jiménez A, Sánchez-Alonso S, Fernández-Díaz C, Alcaraz-Serna A, Mateu-Albero T, Rodríguez-Cortes P, Sánchez-Cerrillo I, et al. IL-6 serum levels predict severity and response to tocilizumab in COVID-19: an observational study. J Allergy Clin Immunol. 2021;147(1):72-80.e78.
Zhao Y, Qin L, Zhang P, Li K, Liang L, Sun J, Xu B, Dai Y, Li X, Zhang C, et al. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight. 2020. https://doi.org/10.1172/jci.insight.139834.
Sweeney JM, Barouqa M, Krause GJ, Gonzalez-Lugo JD, Rahman S, Gil MR. Low ADAMTS13 activity correlates with increased mortality in COVID-19 patients. TH Open. 2021;5(1):e89–103.
Kong Y, Han J, Wu X, Zeng H, Liu J, Zhang H. VEGF-D: a novel biomarker for detection of COVID-19 progression. Crit Care. 2020;24(1):373.
Ichimura T, Mori Y, Aschauer P, Das Padmanabha KM, Padera RF, Weins A, Nasr ML, Bonventre JV. KIM-1/TIM-1 is a receptor for SARS-CoV-2 in lung and kidney. MedRxiv. 2020. https://doi.org/10.1101/2020.09.16.20190694.
Ni W, Yang X, Yang D, Bao J, Li R, Xiao Y, Hou C, Wang H, Liu J, Yang D, et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit Care. 2020;24(1):422.
Gomes CP, Fernandes DE, Casimiro F, da Mata GF, Passos MT, Varela P, Mastroianni-Kirsztajn G, Pesquero JB. Cathepsin L in COVID-19: from pharmacological evidences to genetics. Front Cell Infect Microbiol. 2020;10: 589505.
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–3.
Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, Wang T, Zhang X, Chen H, Yu H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620–9.
Zhang J, Hao Y, Ou W, Ming F, Liang G, Qian Y, Cai Q, Dong S, Hu S, Wang W, et al. Serum interleukin-6 is an indicator for severity in 901 patients with SARS-CoV-2 infection: a cohort study. J Transl Med. 2020;18(1):406.
Zhang J-J, Dong X, Cao Y-Y, Yuan Y-D, Yang Y-B, Yan Y-Q, Akdis CA, Gao Y-D. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75(7):1730–41.
Villar J, Ferrando C, Martinez D, Ambros A, Munoz T, Soler JA, Aguilar G, Alba F, Gonzalez-Higueras E, Conesa LA, et al. Dexamethasone treatment for the acute respiratory distress syndrome: a multicentre, randomised controlled trial. Lancet Respir Med. 2020;8(3):267–76.
Interleukin-6 receptor antagonists in critically Ill patients with covid-19. N Engl J Med 2021, 384(16):1491–1502.
Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18(4):844–7.
George JN, Nester CM. Syndromes of thrombotic microangiopathy. N Engl J Med. 2014;371(7):654–66.
Hariri LP, North CM, Shih AR, Israel RA, Maley JH, Villalba JA, Vinarsky V, Rubin J, Okin DA, Sclafani A, et al. Lung histopathology in coronavirus disease 2019 as compared with severe acute respiratory sydrome and H1N1 Influenza: A systematic review. Chest. 2021;159(1):73–84.
Zhang F, Li W, Feng J, et al. SARS-CoV-2 pseudovirus infectivity and expression of viral entry-related factors ACE2, TMPRSS2, Kim-1, and NRP-1 in human cells from the respiratory, urinary, digestive, reproductive, and immune systems. J Med Virol. 2021;93(12):6671–85.
Narula S, Yusuf S, Chong M, Ramasundarahettige C, Rangarajan S, Bangdiwala SI, van Eikels M, Leineweber K, Wu A, Pigeyre M, et al. Plasma ACE2 and risk of death or cardiometabolic diseases: a case-cohort analysis. Lancet. 2020;396(10256):968–76.
Fagyas M, Fejes Z, Sütő R, Nagy Z, Székely B, Pócsi M, Ivády G, Bíró E, Bekő G, Nagy A, et al. Circulating ACE2 activity predicts mortality and disease severity in hospitalized COVID-19 patients. Int J Infect Dis. 2021. https://doi.org/10.1016/j.ijid.2021.11.028.
Horowitz JE, Kosmicki JA, Damask A, Sharma D, Roberts GHL, Justice AE, Banerjee N, Coignet MV, Yadav A, Leader JB, et al. Genome-wide analysis provides genetic evidence that ACE2 influences COVID-19 risk and yields risk scores associated with severe disease. Nat Genet. 2022. https://doi.org/10.1101/2020.12.14.20248176.
Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23.
Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–20.
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020. https://doi.org/10.1016/S0140-6736(20)30566-3.
Xu XW, Wu XX, Jiang XG, Xu KJ, Ying LJ, Ma CL, Li SB, Wang HY, Zhang S, Gao HN, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368: m606.
Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020. https://doi.org/10.1007/s00134-020-05991-x.
Du R-H, Liang L-R, Yang C-Q, Wang W, Cao T-Z, Li M, Guo G-Y, Du J, Zheng C-L, Zhu Q, et al. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur Respir J. 2020. https://doi.org/10.1183/13993003.00524-2020.
Cummings MJ, Baldwin MR, Abrams D, Jacobson SD, Meyer BJ, Balough EM, Aaron JG, Claassen J, Rabbani LE, Hastie J, et al. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet (London, England). 2020;395(10239):1763–70.
Fang C, Bai S, Chen Q, Zhou Y, Xia L, Qin L, Gong S, Xie X, Zhou C, Tu D, et al. Deep learning for predicting COVID-19 malignant progression. Med Image Anal. 2021;72:102096–102096.
Zhao X, Zhang B, Li P, Ma C, Gu J, Hou P, Guo Z, Wu H, Bai Y. Incidence, clinical characteristics and prognostic factor of patients with COVID-19: a systematic review and meta-analysis. Infect Dis. 2020. https://doi.org/10.1101/2020.03.17.20037572v1.
Juge-Aubry CE, Somm E, Giusti V, Pernin A, Chicheportiche R, Verdumo C, Rohner-Jeanrenaud F, Burger D, Dayer JM, Meier CA. Adipose tissue is a major source of interleukin-1 receptor antagonist: upregulation in obesity and inflammation. Diabetes. 2003;52(5):1104–10.
Group RC, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N, Brightling C, Ustianowski A, et al. Dexamethasone in hospitalized patients with covid-19. N Engl J Med. 2021;384(8):693–704.
Ong SWX, Chiew CJ, Ang LW, Mak TM, Cui L, Toh M, Lim YD, Lee PH, Lee TH, Chia PY, et al. Clinical and virological features of SARS-CoV-2 variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta). Clin Infect Dis. 2021. https://doi.org/10.2139/ssrn.3861566.
Messner CB, Demichev V, Wendisch D, Michalick L, White M, Freiwald A, Textoris-Taube K, Vernardis SI, Egger AS, Kreidl M, et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020;11(1):11-24.e14.
Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C, Quan S, Zhang F, Sun R, Qian L, et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell. 2020;182(1):59-72.e15.
Filbin MR, Mehta A, Schneider AM, Kays KR, Guess JR, Gentili M, Fenyves BG, Charland NC, Gonye ALK, Gushterova I, et al. Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep Med. 2021;2(5): 100287.
Acknowledgements
No further acknowledgements to add.
Funding
The project was supported by American Diabetes Association (https://diabetes.org/), grant number 7–20-COVID-003, awarded to AL. AL was supported by grant 2020096 from the Doris Duke Charitable Foundation (https://www.ddcf.org). RCL, RCC, and ETR were supported by the Centers for Disease Control and Prevention (https://www.cdc.gov), grant numbers U01CK000490 and U01CK000633. LNB was supported by the National Institutes of Health (NIH; https://www.nih.gov), grant number K23DK125839. SJC was supported by the NIH, grant numbers T32DK007028 and F32DK127545. JBM was supported by the NIH, grant numbers R01HL151855 and UM1DK078616. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed significantly to this manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Study procedures were approved by the Mass General Brigham (formerly Partners) Human Research Committee, the governing institutional review board at Massachusetts General Hospital.
Consent for publication
All authors consent to this publication.
Competing interests
SJC reports employment of a family member by a Johnson & Johnson company. The rest of the authors have no potential conflict of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Additional figures and tables.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Schroeder, P.H., Brenner, L.N., Kaur, V. et al. Proteomic analysis of cardiometabolic biomarkers and predictive modeling of severe outcomes in patients hospitalized with COVID-19. Cardiovasc Diabetol 21, 136 (2022). https://doi.org/10.1186/s12933-022-01569-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12933-022-01569-7