Abstract
Background
Recent studies have revealed that serpin peptidase inhibitor clade E member 2 (SERPINE2) is associated with tumorigenesis. However, SERPINE2 expression and its role in lung adenocarcinomas are still unknown.
Methods
The expression levels of SERPINE2 in 74 consecutively resected lung adenocarcinomas were analyzed by using immunostaining. Inhibition of SERPINE2 expression by small interfering RNA (siRNA) was detected by quantitative PCR. Cell number assays and cell apoptosis assays were performed to clarify the cell-autonomous function of SERPINE2 in A549 and PC9 lung cancer cells.
Results
The overall survival of patients with high SERPINE2 expression was significantly worse than that of patients with low SERPINE2 expression (P = 0.0172). Multivariate analysis revealed that SERPINE2 expression was an independent factor associated with poor prognosis (P = 0.03237). The interference of SERPINE2 decreased cell number and increased apoptosis in A549 and PC9 cells
Conclusion
These results suggest that SERPINE2 can be used as a novel prognostic marker of lung adenocarcinoma.
Similar content being viewed by others
Background
Serine proteinase inhibitor clade E member 2 (SERPINE2), also known as protease nexin-1 (PN-1), was first identified as a neurite-promoting factor released by cultured glioma cells [1]. In addition to glioma cells, various other cells secrete SERPINE2, including endothelial cells, fibroblasts, macrophages, platelets, smooth muscle cells, chondrocytes, astrocytes, and several types of tumor cells [2,3,4,5,6]. SERPINE2 was proven to be a member of the SERPINE family, which has serine protease activity [1, 7]. SERPINE2 is overexpressed in a variety of adenocarcinomas, including breast cancer [3], pancreatic cancer [8], gastric cancer [9], and colorectal cancer [10], and its high expression is correlated with the degree of cancer malignancy. A previous study demonstrated that SERPINE2 is upregulated by oncogenic activation of RAS, BRAF and MEK1 and contributes to pro-neoplastic actions of ERK signaling in intestinal epithelial cells [10]. Therefore, SERPINE2 may be a potential therapeutic target for colorectal cancer treatment [10]. In lung adenocarcinomas in particular, high expression of SERPINE2 has been previously reported [11], but the relationship to prognosis or disease progression has never been reported. In this study, we examined the expression of SERPINE2 in 74 consecutive lung adenocarcinoma cases by immunohistochemistry using an anti-SERPINE2 antibody. In addition, we silenced SERPINE2 in two kinds of non-small cell lung cancer (NSCLC) cell lines using siRNA and performed a cell number assay and evaluation of apoptosis to clarify the cell-autonomous function of SERPINE2 in lung adenocarcinoma cell lines.
Methods
Patients
During the period from January 2014 to December 2014, 74 consecutive patients were treated by complete surgical resection of lung adenocarcinoma at Kobe University Hospital, Kobe, Japan. The methods of data collection and analysis were approved by the institutional review board (permission number: 160117), and written informed consent was obtained from all patients.
Construction of the spiral array block and pathological Studies
All surgical specimens were fixed with 10% formalin and embedded in paraffin. The paraffin-embedded block was sent to the Pathology Institute (Toyama, Japan) and processed into a spiral array block. The method of preparing the spiral array block is described in detail elsewhere [12]. Briefly, 50- to 100-μm-thick slices of the sample block are cut and rolled up into cylindrical reels. These cylindrical reels are divided, and the reel containing the target site is embedded vertically in the recipient block. After that, the spiral array block is sliced and used for pathological examination. Serial 4-μm sections were stained with hematoxylin and eosin (Fig. 1a, b). All histologic specimens that had been initially evaluated by pathologists were reviewed, and the expression levels of SERPINE2 were assessed independently by two pathologists (N.J. and T.N.) who were blinded to the clinical data. The histological diagnoses were based on the 2015 WHO classification [13]. Pathological stage was determined on the basis of the TNM classification of the International Union against Cancer (UICC) [14].
Immunohistochemical staining
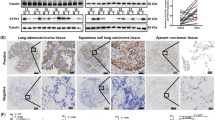
Anti-human SERPINE2 antibody (Lot no. 00014298, Proteintech, Rosemont, IL, USA) was used as the primary antibody. The spiral array block was cut into 4 μm sections, which were mounted on silane-coated slides. The sections were deparaffinized in limonene (Nacalai, Kyoto, Japan) and dehydrated in a graded ethanol series. For antigen retrieval, the slides were heated for 20 min at 121 °C in 10 µM citrate buffer (pH 6.0), and endogenous peroxidase was blocked with 3% hydrogen peroxide in absolute methyl alcohol. The slides were then blocked in 2.5% horse serum for 1 h and incubated with the primary antibodies (1:200). After overnight incubation, the slides were then washed with phosphate–buffered saline and incubated with ImmPRESS Reagent (Vector laboratories, Burlingame, CA, USA). The reaction products were stained with ImmPACT DAB reagent (Vector laboratories), and the sections were counterstained with hematoxylin. A high SERPINE2 case was defined as one in which there was at least a 50% increase in positive cells compared to that in normal tissue (Fig. 1c–f).
Cell culture
The NSCLC cell lines H460, A549 and PC9 were obtained from the American Type Culture Collection (Manassas, VA, USA). All cells were cultured in RPMI 1640 medium (Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin (Wako, Osaka, Japan) under 5% CO2 at 37 °C.
SERPINE2 knockdown by small interfering RNAs (siRNAs)
The SERPINE2 siRNAs (#1: s22188 and #2 s22189) and control siRNA (#14390843) were obtained from Thermo Fisher Scientific, MA, USA. Cells were plated in six-well plates at a density of 2 × 105 cells per well. The siRNAs or control siRNA duplexes were mixed with Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific) in Opti-MEM medium (Thermo Fisher Scientific) as described by the manufacturer’s protocol and added to the plated cells. The cells were used for assessments 24 h after the addition of fresh medium.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Preparation of total cellular RNAs and qRT-PCR of the RNAs by using them as a template were performed as described previously [15]. Relative mRNA levels were calculated with the ∆∆Ct method [16] using GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA as an internal control. The primers used in this study were as follows: 5′-AATGAAACCAGGGATATGATTGAC-3′ and 5′-TTGCAAGATATGAGAAACATGGAG-3′ for SERPINE2 and 5′-GCACCGTCAAGGCTGAGAAC-3′ and 5′-ATGGTGGTGAAGACGCCAGT-3′ for GAPDH.
Effects of SERPINE2 in cell number
To determine the knockdown effects of SERPINE2 on cell growth, we used Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions. A549 and PC9 cells were plated at a concentration of 5 × 103 cells/well in 96-well culture plates and analyzed 72 h later.
Cell migration assay and cell invasion assay
To determine the knockdown effects of SERPINE2 on cell migration and invasion, we used Oris cell migration assay kit and Oris 3D invasion assay kit (Platypus technologies, WI, USA) according to the manufacturer’s instructions. A549 and PC9 cells were plated at a concentration of 2 × 104 cells/well in 96-well assay plates and analyzed 0 h, 24 h, 48 h, and 72 h later. The areas of invasion were measured in ImageJ [17] and we calculated relative invasion areas compared to those at 0 h.
Fas ligand stimulation
To investigate the involvement of SERPINE2 in the apoptosis pathway, lung cancer cell lines in which SERPINE2 was knocked down were stimulated with 200 ng/ml soluble Fas ligand (Wako, Osaka, Japan), and the temporal changes in apoptosis-related proteins were evaluated by western blotting (2, 6, 12, 24, and 48 h after Fas ligand stimulation).
Western blotting
Primary antibodies against the following proteins were purchased from Cell Signaling Technology (Denver, MA, USA): β-actin (#4967), Bax (#2772), Bcl-2 (#4223), cleaved caspase 7 (#8438), and cleaved caspase 9 (#9505). To detect SERPINE2, we used the same antibody used for the immunohistochemical analysis at a different dilution of 1:1000. Cells were lysed in Cell Lysis Buffer (Cell Signaling Technology), and total cellular proteins (20 µg) were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), which was followed by western blotting as described previously [18]. The dilution rate of primary antibody was 1:1000, and incubation performed overnight at 4 degrees Celsius. After washing in TBS-T, the membranes were treated with a HRP-conjugated anti rabbit secondary antibody for 1hour at room temperature. The blots were developed using an enhanced chemiluminescence detection kit (Thermo Fisher Scientific Inc., Waltham, MA, USA).
Evidence from TCGA database
To evaluate the relationship between SERPINE2 mRNA expression and the overall survival of lung adenocarcinoma patients, we utilized The Cancer Genome Atlas (TCGA) dataset through the Gene Expression Profiling Interactive Analysis (GEPIA) web server (http://gepia.cancer-pku.cn/) [19]. In lung adenocarcinoma (LUAD) dataset, we defined the patients in the 1st quartile in terms of SERPINE2 mRNA TPM score in tumor tissue as the low SERPINE2 group and those in the 4th quartile as the high SERPINE2 group.
Statistical analysis
All statistical analyses were performed with EZR version 1.37 (Saitama Medical Center, Jichi Medical University; http://www.jichi.ac.jp/saitama-sct/SaitamaHP.files/statmed.html; Kanda, 2018), which is a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria, version 3.4.1) [20]. Differences in patient characteristics between the two groups were tested for significance by the Pearson χ2 test or the Fisher exact test. For the univariate analysis, the cumulative survival was estimated by the Kaplan–Meier method, and differences in variables were calculated by the log-rank test. A multivariate regression analysis was conducted according to the Cox proportional hazards model. All reported P values are 2-sided, and a P value less than 0.05 was considered significant.
Results
Relationship between SERPINE2 expression and clinicopathologic factors
A consecutive series of 74 specimens of completely resected adenocarcinoma of the lung were examined for SERPINE2 expression. SERPINE2 expression in the cytoplasm of the cancer cells was observed in 19 of the 74 cases (26%). The relationship between SERPINE2 expression and clinicopathologic factors is shown in Table 1. SERPINE2 expression was significantly correlated with lymphatic invasion (P = 0.0188), suggesting that lung adenocarcinomas that express SERPINE2 show aggressive features. There was no relationship between SERPINE2 expression and EGFR mutation status (P = 0.416).
Relationship between SERPINE2 expression in immunostaining and overall survival
The overall survival curve obtained by the Kaplan–Meier method is shown in Fig. 2. SERPINE2 expression was significantly correlated with a short survival time (P = 0.0172). Univariate analyses showed that high expression of SERPINE2 (P = 0.03228) was correlated with a short survival time (Table 2). Table 3 shows the impact of potential prognostic factors on the survival of patients with adenocarcinoma with expression of SERPINE2 based on the results of the multivariate analysis with the Cox proportional hazards model. All significant univariate parameters were included in the multivariate analysis simultaneously. Together, these results proved that SERPINE2 was an independent predictor of a poor prognostic outcome.
Survival analysis of TCGA dataset
The survival analysis of the TCGA dataset performed through the GEPIA webserver also showed that the high SERPINE2 group had a worse prognosis (P = 0.042) than the low expression group (Fig. 3).
Effect of SERPINE2 siRNA on the autonomous behavior of cancer cells
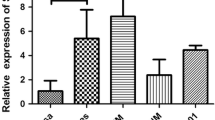
Among lung cancer cell lines, H460, A549 and PC9 more highly express SERPINE2 compared to BEAS-2B, which is a bronchial epithelial cell line (Fig. 4a). To test the effect of increased SERPINE2 on cell autonomous behavior in vitro, the cell number of A549 and PC9 cells treated with SERPINE2 siRNA or a negative control was examined by using a CCK-8 assay. After transfection, the interference efficiency of SERPINE2 was first confirmed by qRT-PCR. Normalized to the expression of the negative control, SERPINE2 expression was significantly downregulated in A549 and PC9 cells by both SERPINE2 siRNA#1 and #2 (p < 0.05) (Fig. 4b). The protein levels of SERPINE2 were also suppressed by gene knockdown (Fig. 4c). Then, the cell number was quantified at 24, 48, and 72 h. The cell number was significantly decreased in cancer cells with SERPINE2 downregulation (P < 0.05) (Fig. 5). This result suggests that SERPINE2 possibly plays a role in the proliferation of cancer cells.
SERPINE2 expression and knockdown effect of SERPINE2. a SERPINE2 expression in lung cancer cell lines was normalized to BEAS-2B. b, c The efficacies of siRNA in A549 and PC9 cells were analyzed by qRT-PCR (B, relative to negative control) and western blotting (c). nc, negative control; #1, siRNA#1; #2, siRNA#2
The effect of SERPINE2 knockdown on Fas ligand-mediated apoptosis
First, we performed cell migration and invasion assays, but we did not observe remarkable changes (Additional file 1: Figs. S1 and S2). Furthermore, BMP4, which plays a crucial role in migration and invasion, was not activated by SERPINE2 knockdown in A549 and PC9 lung cancer cells (Additional file 1: Fig. S3). Then, to investigate the effect of SERPINE2 on apoptosis, A549 and PC9 cells treated with SERPINE2 siRNA or a negative control siRNA were stimulated with soluble Fas ligand (200 ng/ml). In both cell lines, Fas ligand-mediated apoptosis was more highly induced in cells with SERPINE2 interference. Especially in A549 cells, the protein expression of bcl-2, an anti-apoptotic protein, was decreased (Fig. 6). Together, these results suggest that SERPINE2 is not associated with cell migration and invasion but is associated with anti-apoptosis.
Effect of SERPINE2 knockdown on apoptosis. Western blotting analysis of apoptosis-related proteins at the indicated time points after Fas ligand stimulation is shown. A549 (upper) and PC9 (lower) cells pretreated with SERPINE2 siRNA were stimulated with Fas ligand (200 ng/ml). nc, negative control; #2, siRNA#2
Discussion
The current study demonstrated the role of SERPINE2 as an independent prognostic factor of lung adenocarcinomas and its molecular mechanism for the first time. In terms of prognostic factors, evaluation of the pathological specimens showed that increased expression of SERPINE2 was related to lymphatic invasion and poor overall survival. Using the GEPIA database, we found that high SERPINE2 mRNA expression was also related to poor prognosis. The overall results for survival showed the same trend as that observed in previous reports on different cancers, including breast cancers [3], gastric cancers [9] and osteosarcomas [21]. Regarding the molecular mechanism, in vitro analysis revealed that SERPINE2 plays a possible anti-apoptotic role. It may be related to the malignant features in cancer cells with high SERPINE2 expression.
Our experiment showed that silencing of SERPINE2 resulted the decrease in cell number. Epidermal growth factor (EGF) induced SERPINE2 expression through the EGF/MEK/ERK pathway, and SERPINE2 knockdown reduced cell proliferation induced by EGF [22]. SERPINE2 is thought to act as an effector of the EGF pathway to cause cell proliferation. Furthermore, SERPINE2 is reported to inhibit plasminogen-induced apoptosis of Chinese hamster ovary fibroblasts (CHO-K1), which constitutively express tissue-type plasminogen activator (t-PA) [23]. In a previous study, transfection of the SERPINE2 gene significantly inhibited the activity of plasmin and t-PA via the formation of inhibitory complexes and prevented cell detachment and apoptosis [23]. On the other hand, it was reported in a study using prostate cancer cells that apoptosis was induced by SERPINE2 [5], and it is believed that the effect differs depending on the cancer type. The results of the current experiment suggest that SERPINE2 suppresses apoptosis in lung cancer cell lines and may be a target molecule for lung cancer treatment.
In esophageal squamous cell carcinomas, SERPINE2 inhibition resulted in a reduction in cell growth, migration and invasion [24]. In esophageal squamous cell carcinomas, SERPINE2 promotes tumor metastasis by activating bone morphogenetic protein 4 (BMP4) [24]. Hence, we revealed no remarkable changes in cell migration and invasion assays (Additional file 1: Figs. S1 and S2) because the current study revealed that SERPINE2 was not associated with BMP4 in lung cancer cells (Additional file 1: Fig. S3).
The limitation of the current study is that a relatively small sample size is included in the current study. Hence, other prognostic factors, such as T and N factors, were not significant factors associated with poor prognosis.
Conclusions
SERPINE2 can be a prognostic factor and might be a possible target of NSCLC by suppressing apoptosis.
Abbreviations
- BMP4:
-
Bone morphogenetic protein 4
- CCK-8:
-
Cell Counting Kit-8
- NSCLC:
-
Non-small cell lung cancer
- qRT-PCR:
-
Quantitative reverse transcription-polymerase chain reaction
- PN-1:
-
Protease nexin-1
- SDS-PAGE:
-
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SERPINE2:
-
Serine proteinase inhibitor clade E member 2
- siRNA:
-
Small interfering RNA
- UICC:
-
International Union against Cancer
References
Guenther J, Nick H, Monard D. A glia-derived neurite-promoting factor with protease inhibitory activity. EMBO J. 1985;4:1963–6.
Mansilla S, Boulaftali Y, Venisse L, Arocas V, Meilhac O, Michel JB, Jandrot-Perrus M, Bouton MC. Macrophages and platelets are the major source of protease nexin-1 in human atherosclerotic plaque. Arterioscler Thromb Vasc Biol. 2008;28:1844–50.
Fayard B, Bianchi F, Dey J, Moreno E, Djaffer S, Hynes NE, Monard D. The serine protease inhibitor protease nexin-1 controls mammary cancer metastasis through LRP-1-mediated MMP-9 expression. Cancer Res. 2009;69:5690–8.
Rosenblatt DE, Cotman CW, Nieto-Sampedro M, Rowe JW, Knauer DJ. Identification of a protease inhibitor produced by astrocytes that is structurally and functionally homologous to human protease nexin-I. Brain Res. 1987;415:40–8.
McKee CM, Ding Y, Zhou J, Li C, Huang L, Xin X, He J, Allen JE, El-Deiry WS, Cao Y, et al. Protease nexin 1 induces apoptosis of prostate tumor cells through inhibition of X-chromosome-linked inhibitor of apoptosis protein. Oncotarget. 2015;6:3784–96.
Santoro A, Conde J, Scotece M, Abella V, Lois A, Lopez V, Pino J, Gomez R, Gomez-Reino JJ, Gualillo O. SERPINE2 inhibits IL-1α-induced MMP-13 expression in human chondrocytes: involvement of ERK/NF-κB/AP-1 pathways. PLoS ONE. 2015;10:e0135979.
Stone SR, Nick H, Hofsteenge J, Monard D. Glial-derived neurite-promoting factor is a slow-binding inhibitor of trypsin, thrombin, and urokinase. Arch Biochem Biophys. 1987;252:237–44.
Buchholz M, Biebl A, Neesse A, Wagner M, Iwamura T, Leder G, Adler G, Gress TM. SERPINE2 (protease nexin I) promotes extracellular matrix production and local invasion of pancreatic tumors in vivo. Cancer Res. 2003;63:4945–51.
Wang K, Wang B, Xing AY, Xu KS, Li GX, Yu ZH. Prognostic significance of SERPINE2 in gastric cancer and its biological function in SGC7901 cells. J Cancer Res Clin Oncol. 2015;141:805–12.
Bergeron S, Lemieux E, Durand V, Cagnol S, Carrier JC, Lussier JG, Boucher MJ, Rivard N. The serine protease inhibitor serpinE2 is a novel target of ERK signaling involved in human colorectal tumorigenesis. Mol Cancer. 2010;9:271.
Yang Y, Xin X, Fu X, Xu D. Expression pattern of human SERPINE2 in a variety of human tumors. Oncol Lett. 2018;15:4523–30.
Fukuoka J, Hofer MD, Hori T, Tanaka T, Ishizawa S, Nomoto K, Saito M, Uemura T, Chirieac LR. Spiral array: a new high-throughput technology covers tissue heterogeneity. Arch Pathol Lab Med. 2012;136:1377–84.
Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances Since the 2004 classification. J Thorac Oncol. 2015;10:1243–60.
TNM Classification of Malignant Tumours. 7 ed. Wiley-Blackwell; 2011.
Nagano T, Edamatsu H, Kobayashi K, Takenaka N, Yamamoto M, Sasaki N, Nishimura Y, Kataoka T. Phospholipase cε, an effector of ras and rap small GTPases, is required for airway inflammatory response in a mouse model of bronchial asthma. PLoS ONE. 2014;9:e108373.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5.
Hatakeyama Y, Kobayashi K, Nagano T, Tamura D, Yamamoto M, Tachihara M, Kotani Y, Nishimura Y. Synergistic effects of pemetrexed and amrubicin in non-small cell lung cancer cell lines: potential for combination therapy. Cancer Lett. 2014;343:74–9.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–102.
Kanda Y. Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Transplant. 2013;48:452–8.
Mao M, Wang W. SerpinE2 promotes multiple cell proliferation and drug resistance in osteosarcoma. Molecular medicine reports. 2016b;14:881–7.
Tang T, Zhu Q, Li X, Zhu G, Deng S, Wang Y, Ni L, Chen X, Zhang Y, Xia T, et al. Protease nexin I is a feedback regulator of EGF/PKC/MAPK/EGR1 signaling in breast cancer cells metastasis and stemness. Cell Death Dis. 2019;10:649.
Rossignol P, Ho-Tin-Noé B, Vranckx R, Bouton M-C, Meilhac O, Lijnen RH, Guillin M-C, Michel J-B, Anglés-Cano E. Protease nexin-1 inhibits plasminogen activation-induced apoptosis of adherent cells. J Biol Chem. 2004;279:10346–56.
Zhang J, Luo A, Huang F, Gong T, Liu Z. SERPINE2 promotes esophageal squamous cell carcinoma metastasis by activating BMP4. Cancer Lett. 2020;469:390–8.
Acknowledgements
The results shown here are in whole or in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
We thank all the members of the Division of Respiratory Medicine, Kobe University Graduate School of Medicine, for their helpful discussions.
Funding
This work was supported by funding from JSPS KAKENHI 17K09614 and Takeda Science Foundation to Tatsuya Nagano.
Author information
Authors and Affiliations
Contributions
RD and TN wrote the manuscript. RD performed immunostaining. TK and HS made the database. TN and NJ performed pathological analysis. YM collected the samples. YY, MY and KK assisted with technical advice. MT and YN conducted the statistical analysis. All authors analyzed the data, and conceived the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The methods of data collection and analysis were approved by the institutional review board (permission number: 160117), and written informed consent was obtained from all the patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Additional figures.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Dokuni, R., Nagano, T., Jimbo, N. et al. High expression level of serpin peptidase inhibitor clade E member 2 is associated with poor prognosis in lung adenocarcinoma. Respir Res 21, 331 (2020). https://doi.org/10.1186/s12931-020-01597-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12931-020-01597-5