Abstract
Background
Chronic Obstructive Pulmonary Disease (COPD) is associated with an abnormal pulmonary and systemic immune response to tobacco smoking. Yet, how do immune cells relate within and between these two biological compartments, how the pulmonary infiltrate influences the lung transcriptome, and what is the role of active smoking vs. presence of disease is unclear.
Methods
To investigate these questions, we simultaneously collected lung tissue and blood from 65 individuals stratified by smoking habit and presence of the disease. The immune cell composition of both tissues was assessed by flow cytometry, whole lung transcriptome was determined with Affymetrix arrays, and we used Weighted Gene Co-expression Network Analysis (WGCNA) to integrate results.
Results
Main results showed that: (1) current smoking and the presence of COPD were both independently associated with a reduction in the proportion of lung T cells and an increase of macrophages, specifically those expressing CD80 + CD163+; (2) changes in the proportion of infiltrating macrophages, smoking status or the level of airflow limitation were associated to different WGCNA modules, which were enriched in iron ion transport, extracellular matrix and cilium organization gene ontologies; and, (3) circulating white blood cells counts were correlated with lung macrophages and T cells.
Conclusions
Mild-moderated COPD lung immune infiltrate is associated with the active smoking status and presence of disease; is associated with changes in whole lung tissue transcriptome and marginally reflected in blood.
Similar content being viewed by others
Background
Tobacco smoking is the main environmental risk factor for COPD, albeit not all smokers develop the disease [1, 2]. It is currently accepted that, in so-called “susceptible smokers”, tobacco smoking triggers an abnormal pulmonary and systemic immune response that eventually damages the lung parenchyma and leads to persistent airflow limitation and COPD [1,2,3,4,5]. Yet, some aspects of this pathogenic paradigm are still unclear and require specific research. For instance, although in cross-sectional studies [6,7,8,9] the number of inflammatory cells infiltrating the lung parenchyma increase in parallel with the severity of airflow limitation, how do these cells relate among themselves is unknown. Likewise, it is unclear how changes in the pulmonary immune cell network relate to changes in the whole lung transcriptome [10,11,12,13]. Besides, given that smoking per se has well recognized effects on the immune system [4, 14], it is important (and challenging) to disentangle the role of disease itself vs. that of active smoking. Finally, previous studies have tried to identify blood surrogate markers of lung changes [15, 16], but the relation between different immune cell types in the two compartments (i.e. lung and blood) has not been ever assessed.
We hypothesized that a data set that combined lung tissue and circulating blood measurements collected simultaneously in current and former smokers with COPD, as well as controls with normal lung function (current and never smokers), would be a unique asset to: (1) characterize the lung and blood immune cell composition in relation to both smoking status and presence of disease; (2) explore the relationship between the lung immune cell composition and the lung tissue transcriptome; and, finally, (3) construct a multi-level (lung and blood) immune cell correlation network to assess if the lung changes are reflected in the blood.
Methods
Full methods are provided in the Additional file 1.
Population and ethics
We prospectively collected clinically relevant information as well as lung tissue and blood samples from 65 patients who required thoracic surgery for lung cancer (Table 1). The lung tissue was from a non-affected location. All participants signed their informed consent, and the Ethics Committee of our institution approved the study (HCB-2012/7731).
Lung tissue processing
Fresh lung tissue was washed with PBS, enzymatically digested with 0.5 mg/ml Collagenase P and 0.1 mg/ml DNase I (Roche, Germany), incubated at 37 °C for 30 min, and mechanically digested with GentleMACS M-dissociator tubes (Miltenyi Biotech, Germany).
Measurements
Lung function
Forced spirometry and carbon monoxide lung capacity (DLCO) were determined according to international recommendations [2] and reference values corresponded to a Mediterranean population [17].
Circulating inflammatory markers
Circulating venous blood was collected in EDTA tubes (BD, US) in the operating room before surgery started. Total white blood cells counts were quantified with an ADVIA-2120 system (Siemens, Germany). The plasma concentration of high sensitivity C Reactive Protein (hsCRP) was determined by ultra-sensitive quantitative turbidimetric test (Bayer Diagnostics, Germany).
Flow cytometry
Immune cell populations were analysed in parallel in blood and lung tissue. Lung tissue homogenates (5·105 cells) or 50 μl of peripheral blood were stained with the corresponding fluorescently conjugate monoclonal antibody mix (Additional file 1: Table S1), incubated at 4 °C for 30 min and washed. A minimum of 2·105 cells per tube were acquired in a FacsCanto (BD Biosciences, US), data was analyzed using FlowJo v10 software (FlowJo LLC, US) Additional file 2: Figures S1-S6.
Transcriptomic analysis
Total RNA was isolated with PureLink RNA-MiniKit (Life Technologies, US), quantified by Nanodrop (Thermo Scientific, Germany). RNA samples with integrity numbers (RIN) ≥ 7 (Agilent technologies, Germany), where analysed with the Affymetrix GeneChip® Human Genome U219 Array Plate. Only from 53 individuals the mRNA quality was adequate for analysis (Additional file 1: Table S2). Raw microarray data has been deposited in GEO (GSE103174).
Data analysis
Statistical analysis were performed using R [18]. Results are presented as n, proportion, or mean ± standard deviation. Differences between groups were assessed using the Kruskal-Wallis test, followed by post-hoc contrasts if necessary (Table 2). A linear model after log transformation of variables was used to ascertain the independent contribution of smoking status and presence of disease. P values < 0.05 were considered significant. Lung-blood immune cell correlation networks were built with R and visualized using Cytoscape as described elsewhere (Spearman coefficient r > |0.3| and a p-value < 0.05) [19].
Results
Participant characteristics
Table 1 presents the main characteristics of participants. In patients with COPD the proportion of subjects with GOLD grade 1 airflow limitation (31% (n = 5) vs. 32% (n = 9)) and GOLD grade 2 (69% (n = 11) vs. 68% (n = 19)) was similar in former (FS) and current smokers (CS) patients, respectively. DLCO was mildly reduced in COPD patients. Circulating leukocyte and neutrophil counts and hsCRP plasma levels were increased in current smokers (with or without COPD) (Table 1).
Characterization of lung and blood immune cells
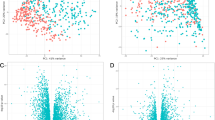
Figure 1 (panels a and b) compares the proportion of the different lung immune cell populations studied, expressed as percentage of total CD45+ cells (hematopoietic lineage, a common practice in order to perform immunophenotyping in non-lymphoid tissues [20]) in the four groups investigated. Main differences were observed in current smokers with COPD (COPD-CS) who showed: (1) a significant reduction in the proportion of T-cells that involved both CD4+ and CD8+ T cells (Table 2 and Additional file 2: Figure S7); (2) a significant increase in the proportion of macrophages (Mϕ) (Fig. 1a), of whom a higher percentage expressed both CD80 + CD163+ (Table 2, Additional file 2: Figure S8) and a relative reduction of CD80 + CD163-Mϕ (Table 2, Additional file 2: Figure S8); and, (3) a different distribution of lung monocytes between study groups (Fig. 1b) due to an increase of monocytes in COPD-CS (Table 2, Additional file 2: Figure S8D).
Proportion of immune cell populations (mean ± SD), expressed as percentage of total CD45+ cells, both in lung tissue (Top panels a and b) and circulating blood (Bottom panels c and d). Study groups: never smokers (white bars), smokers with normal lung function (grey bars), COPD-former smokers (COPD-FS, gray stripped bars) and COPD current smokers (COPD-CS, black bars). Note the different Y scales in right vs. left panels. For further explanations, see text
As main changes were observed in COPD-CS, next we used a linear model in order to evaluate if both current smoking and presence of COPD have an independent effect. We observed that current smoking was inversely related to the proportion of lung T-lymphocytes (βeta = − 0.111, p value =0.008) and positively with that of macrophages (Mϕ) (βeta =0.210, p value =0.001) and monocytes (βeta =0.104, p value =0.005); while the presence of COPD had an independent but smaller effect (Mϕ, βeta =0.152, p value =0.05; T-lymphocytes, βeta = − 0.100, p value =0.065).
Finally, the proportion of the different immune cell populations in circulating blood was not different across groups (Fig. 1c and d).
Relationship between lung immune cell population and whole lung transcriptome
To assess the association between the immune composition of the lung and the biological processes on-going in the lung, we measured the gene expression profile of the whole lung from 53 participants (82% of the total population studied, Additional file 1: Table S2) and used Weighted Gene Co-expression Network Analysis (WGCNA) to integrate the information. WGCNA defines a set of modules of co-expressed genes, and for each provides their “eigengene” value (i.e., the principal component of the expression matrix of the probes within the module). The eigengene can be related to the variables of interest [21], in our case FEV1, the smoking status and the percentage of lung macrophages (the cell population reporting more differences across study groups).
WGCNA identified 23 modules (Additional file 1: Table S3), of them in eight the eigengene was significantly associated with the variables of interest (Fig. 2).
Heat-map of module association with: the level of FEV1% predicted, the smoking status, and the percentage of Macrophages (Mϕ)). The top number in each cell corresponds to the effect from the linear regression and the bottom number is the p-value. In the box, the summarizing terms for the Gene Ontology (GO) terms significantly enriched for this module obtained from the Revigo analysis are displayed. The full list of GO terms if provided in Additional file 1: Table S5. For further explanations, see text
Specifically, we observed that one module (Yellow, n = 645 genes) was associated to the levels of FEV1 and DLCO % predicted. (Fig. 3) and was enriched in genes associated to the Cilium Organization Gene Ontology biological process term (GO) (Additional file 1: Tables S4-S5).
Multilevel (lung and blood) Spearman correlation network (|r| > 0.3, p < 0.05) for all participants (n = 65) of lung (green), peripheral blood (red) immune cell populations, lung function variables (blue) and general systemic inflamatory biomarkers (yellow). Positive correlations are represented with a continuous edge while the negatives are with a discontinuous edge. Edge witdth is proportional to the strength of the correlation. Node size is proportional to the mean percentatge of CD45% cells of the population. Blue links are used to highlight correlations between lung and blood markers. For further explanations, see text
Three modules were associated both to the smoking status and percentage of infiltrating Mϕ (Fig. 3): (i) Tan (n = 211), enriched for iron metabolism and phospholipid metabolism GO terms (Additional file 1: Tables S4-S5), (iii) Royalblue (n = 86), related to T cell activation GO terms (Additional file 1: Tables S4-S5), (iii) Lightcyan (n = 140), related to mRNA processing GO terms (Additional file 1: Tables S4-S5).
Three modules were associated to the level of lung Mϕ expressing different surface markers (Fig. 2 and Additional file 2: Figure S9): i) Greenyellow (n = 227), especially related to CD80 + CD163+ Mϕ, with genes enriched in extracellular matrix organization and collagen metabolism GO terms (Additional file 1: Tables S4-S5). Interestingly this module also presented a negative correlation with the levels of DLCO % ref. (ii) Blue (n = 789), related to CD80-CD163+ Mϕ, with genes enriched in ER to Golgi vesicle transport and regulation of translation GO terms (S4-S5). (iii) LightCyan (n = 140), related to CD80-CD163+ Mϕ, with genes enriched in mRNA processing (S4-S5).
Finally, we assessed which of the module genes have been previously identified as differentially expressed in lung tissue of COPD (GSE47460). The 10 genes with highest differential expression (DE) included are reported in Table 3. Interestingly, the modules including more differentially expressed genes were the GreenYellow (n = 50) and the Yellow (n = 25) (Additional file 1: Table S6).
Multi-level (blood and lung) immune cell correlation network
Finally, we build a multi-level correlation network [22] to investigate the potential relationships between immune cells and clinical variables in both lung and blood (Fig. 3) and observed that: (1) the strongest correlations were found between cells of the same tissue (percentage of lung Mϕ and T cells (r = − 0.81) and percentage of blood T cells and neutrophils (r = − 0.73); (2) eight of the 40 total connections (20%) involved lung and blood populations (Fig. 2, highlighted with blue edges). Of note, that the percentage of lung T cells was related to the percentage of blood T cells (r = 0.34), and the absolute counts of leukocytes (r = − 0.33). Interestingly, the absolute counts of blood leukocytes was also positively correlated with the proportion of lung Mϕ (r = 0.3); (3) FEV1% predicted was positively correlated with the proportion of lung T cells as well as lung and blood NKT cells, whereas it was negatively correlated with the proportion of lung DCs; and, finally (4) Carbon monoxide lung diffusing capacity (DLCO, % ref.), a well-validated surrogate marker of emphysema [23], was negatively correlated with the proportion of lung Mϕ, DCs and Monocytes, and positively correlated with those of lung T cells and peripheral blood T cells. It is of note that clinical variables were mostly correlated with lung tissue (not blood) populations (Fig. 3).
Discussion
The main and novel observations of this study are that: (1) the composition of the immune cell infiltrate in the lung (but not in circulating blood) is significantly altered by active smoking and the presence of mild-moderate COPD. These changes are characterized by a reduction in the proportion of lung CD4+ and CD8+ T lymphocytes, and an increase in that of Mϕ, especially those expressing CD80 and CD163; (2) the percentage of lung tissue Mϕ is associated, independently of the airflow limitation and smoking status, to several modules of co-expressed genes in whole lung tissue including genes that have been previously associated with the pathobiology of COPD; and, finally (3) the immune cell multi-level correlation network shows some weak relations between lung and blood cells, the strongest one between the absolute number of blood leukocytes and the proportion of lung macrophages and T cells.
Many previous studies have investigated the pulmonary and systemic inflammatory response in COPD [6,7,8]. It is generally accepted that the absolute number of T-cells, both CD8+ cytotoxic T lymphocytes [24] and CD4+ T lymphocytes (polarized towards a TH1 response) [25], increase in patients with severe-very severe COPD. Our results in lung tissue show a reduction in the proportion of infiltrating T cells which was mostly related to smoking and only marginally to the presence of COPD. Apparently, therefore, our results do not agree with previous reports. However, to interpret them appropriately, it is important to consider that: (1) because our methodology does not allow the quantification of the absolute number of infiltrating cells, our results refer to their relative proportion (not to their absolute numbers); (2) we studied patients with mild-moderate, not severe-very severe, COPD. In fact, a recent study in bronchoalveolar lavage fluid (BALF) obtained from patients with mild-moderate COPD (GOLD grades 1–2) also reported a decrease in the proportion of T CD4+ cells and a clear effect of active smoking on cell population distribution [26]. This is indeed in line with our results and highlighting that GOLD grade and current smoking have both to be carefully considered when investigating the inflammatory response of COPD. On the other hand, it is well established that Mϕ are increased in BALF and lung tissue of COPD patients [27]. Our results are in keeping with these previous observations and extend them by showing that this increase is related independently both to active smoking and, to a lesser extent, to the presence of the disease.
Several previous studies have investigated lung transcriptomics in COPD [28, 29], but to our knowledge none has previously related the transcriptomic status with the proportion of infiltrating macrophages. Using WGCNA we identified four transcriptomic co-expression modules that were associated with the proportion of lung Mϕ independently of the level of airflow limitation and smoking status. Interestingly one module (GreenYellow) was associated with the percentage of CD80 + CD163+ macrophages and enriched in ontologies related to extracellular matrix organization and collagen metabolism. Interestingly this module presented a negative correlation with the levels of DLCO, in agreement with the findings of the multi-level correlation network. This module included genes differentially expressed in other lung tissue cohorts as: COL14A1, SFRP2 and CCDC80. Several reports have identified a differential expression of collagen genes in COPD [10], while SFRP2 has been related to a down-regulation of the WNT pathway and CCDC80 is expressed by alveolar macrophages [30]. Although this is a starting point for future research aimed at understanding mechanistically the pathogenic relevance of these associations, is in line with our previous reports of an abnormal catabasis in lung tissue macrophages of patients with COPD [31].
Finally, previous studies reported alterations in specific subsets of circulating blood cells, specifically an increase in total leukocytes and neutrophils in relation to current smoking has been described [5, 32]. But none used network analysis to relate lung and blood immune cells with clinical characteristics. Our results showed that circulating white cells (both total leukocyte counts) were negatively correlated with the percentage of lung T-cells and positively correlated with the percentage of lung Mϕ. Yet, the overall relation between the two compartments (lung and blood) is weak, so the blood immune cell composition cannot be used to predict the inflammatory status of the lung tissue.
The strengths of the study are that, for the first time it profiles a wide variety of immune cells, both in lung tissue and circulating blood, of a relatively large population of patients with mild moderated COPD and controls and uses multi-level network analysis to relate the immune response of the pulmonary and systemic compartments. Likewise, it is the first to investigate the lung transcriptome with the immune cell infiltrate. On the other hand, the limitations of the study are that all participants had surgically resectable lung cancer and that, although analysed lung samples were tumor-free, this can be a potentially confounding factor in our analysis. However, if this was the case, it might have affected similarly all samples analysed. Finally, it would have been interesting to assess how the immune infiltrate changes according to both current smoking and inhaled corticosteroid treatment (ICS) treatment, yet our cohort is underpowered to do these analysis as only COPD 8 patients were under ICS treatment.
Conclusions
This study, by integrating high throughput analytical techniques in a unique, multi-level data set, provides novel information on the relation of the pulmonary and systemic inflammatory response that characterizes COPD. Our main findings highlight the role of lung macrophages and the active smoking status in the pathobiology of COPD. All in all, these observations contribute to a better understanding of COPD pathobiology.
Availability of data and materials
Data is available from GEO#(GSE103174) and from authors on a reasonable request.
Abbreviations
- BMI:
-
Body mass index
- COPD:
-
Chronic obstructive pulmonary disease
- DC:
-
Dendritic cells
- Mϕ:
-
Macrophages
- NKT:
-
Natural killer T cells
- WGCNA:
-
Weighted Gene Co-expression Network Analysis
References
Faner R, Cruz T, Agusti A. Immune response in chronic obstructive pulmonary disease. Expert Rev Clin Immunol. 2013;9:821–33.
Vogelmeier CF, Criner GJ, Martinez FJ, Anzueto A, Barnes PJ, Bourbeau J, Celli BR, Chen R, Decramer M, Fabbri LM, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. GOLD executive summary. Am J Respir Crit Care Med. 2017;195:557–82.
Hodge G, Mukaro V, Reynolds PN, Hodge S. Role of increased CD8/CD28(null) T cells and alternative co-stimulatory molecules in chronic obstructive pulmonary disease. Clin Exp Immunol. 2011;166:94–102.
Faner R, Gonzalez N, Cruz T, Kalko SG, Agusti A. Systemic inflammatory response to smoking in chronic obstructive pulmonary disease: evidence of a gender effect. PLoS One. 2014;9:e97491.
Agusti A, Edwards LD, Rennard SI, MacNee W, Tal-Singer R, Miller BE, Vestbo J, Lomas DA, Calverley PM, Wouters E, et al. Persistent systemic inflammation is associated with poor clinical outcomes in COPD: a novel phenotype. PLoS One. 2012;7:e37483.
Hogg JC, Chu F, Utokaparch S, Woods R, Elliott WM, Buzatu L, Cherniack RM, Rogers RM, Sciurba FC, Coxson HO, Pare PD. The nature of small-airway obstruction in chronic obstructive pulmonary disease. NEnglJMed. 2004;350:2645–53.
Cosio MG, Saetta M, Agusti A. Immunologic aspects of chronic obstructive pulmonary disease. NEnglJMed. 2009;360:2445–54.
Brusselle GG, Joos GF, Bracke KR. New insights into the immunology of chronic obstructive pulmonary disease. Lancet. 2011;378:1015–26.
Van Pottelberge GR, Bracke KR, Van den Broeck S, Reinartz SM, van Drunen CM, Wouters EF, Verleden GM, Vermassen FE, Joos GF, Brusselle GG. Plasmacytoid dendritic cells in pulmonary lymphoid follicles of patients with COPD. Eur Respir J. 2010;36:781–91.
Faner R, Cruz T, Casserras T, Lopez-Giraldo A, Noell G, Coca I, Tal-Singer R, Miller B, Rodriguez-Roisin R, Spira A, et al. Network analysis of lung transcriptomics reveals a distinct B cell signature in emphysema. Am J Respir Crit Care Med. 2016;193(11):1242–53.
Morrow JD, Zhou X, Lao T, Jiang Z, DeMeo DL, Cho MH, Qiu W, Cloonan S, Pinto-Plata V, Celli B, et al. Functional interactors of three genome-wide association study genes are differentially expressed in severe chronic obstructive pulmonary disease lung tissue. Sci Rep. 2017;7:44232.
Obeidat M, Nie Y, Fishbane N, Li X, Bosse Y, Joubert P, Nickle DC, Hao K, Postma DS, Timens W, et al. Integrative genomics of emphysema associated genes reveals potential disease biomarkers. Am J Respir Cell Mol Biol. 2017;57(4):411–8.
de Vries M, Faiz A, Woldhuis RR, Postma DS, de Jong TV, Sin DD, Bosse Y, Nickle DC, Guryev V, Timens W, et al. Lung tissue gene-expression signature for the ageing lung in COPD. Thorax. 2017;73(7). https://doi.org/10.1136/thoraxjnl-2017-210074.
Stampfli MR, Anderson GP. How cigarette smoke skews immune responses to promote infection, lung disease and cancer. Nat Rev Immunol. 2009;9:377–84.
Barnes PJ, Celli BR. Systemic manifestations and comorbidities of COPD. Eur Respir J. 2009;33:1165–85.
Agusti A, Gea J, Faner R. Biomarkers, the control panel and personalized COPD medicine. Respirology. 2016;21:24–33.
Roca J, Sanchis J, Agusti-Vidal A, Segarra F, Navajas D, Rodriguez-Roisin R, Casan P, Sans S. Spirometric reference values from a Mediterranean population. Bull Eur Physiopathol Respir. 1986;22:217–24.
Computing. RCTRFfS: R: A language and environment for statistical computing. 2016.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Yu YR, O'Koren EG, Hotten DF, Kan MJ, Kopin D, Nelson ER, Que L, Gunn MD. A protocol for the comprehensive flow cytometric analysis of immune cells in Normal and inflamed murine non-lymphoid tissues. PLoS One. 2016;11:e0150606.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559.
Noell G, Cosio BG, Faner R, Monso E, Peces-Barba G, de Diego A, Esteban C, Gea J, Rodriguez-Roisin R, Garcia-Nunez M, et al. Multi-level differential network analysis of COPD exacerbations. Eur Respir J. 2017;50(3). https://doi.org/10.1183/13993003.00075-2017.
Barjaktarevic I, Springmeyer S, Gonzalez X, Sirokman W, Coxson HO, Cooper CB. Diffusing capacity for carbon monoxide correlates best with tissue volume from quantitative CT scanning analysis. Chest. 2015;147:1485–93.
Saetta M, Di Stefano A, Turato G, Facchini FM, Corbino L, Mapp CE, Maestrelli P, Ciaccia A, Fabbri LM. CD8+ T-lymphocytes in peripheral airways of smokers with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1998;157:822–6.
Turato G, Zuin R, Miniati M, Baraldo S, Rea F, Beghé B, Monti S, Formichi B, Boschetto P, Harari S, et al. Airway inflammation in severe chronic obstructive pulmonary disease: relationship with lung function and radiologic emphysema. Am J Respir Crit Care Med. 2002;166:105–10.
Forsslund H, Mikko M, Karimi R, Grunewald J, Wheelock Å, Wahlström J, Sköld CM. Distribution of T-cell subsets in BAL fluid of patients with mild to moderate COPD depends on current smoking status and not airway obstruction. Chest. 2014;145:711–22.
Molet S, Belleguic C, Lena H, Germain N, Bertrand CP, Shapiro SD, Planquois JM, Delaval P, Lagente V. Increase in macrophage elastase (MMP-12) in lungs from patients with chronic obstructive pulmonary disease. Inflamm Res. 2005;54:31–6.
Gower AC, Steiling K, Brothers JF, Lenburg ME, Spira A. Transcriptomic studies of the airway field of injury associated with smoking-related lung disease. Proc Am Thorac Soc. 2011;8:173–9.
Zeskind JE, Lenburg ME, Spira A. Translating the COPD transcriptome: insights into pathogenesis and tools for clinical management. Proc Am Thorac Soc. 2008;5:834–41.
Poczobutt JM, De S, Yadav VK, Nguyen TT, Li H, Sippel TR, Weiser-Evans MC, Nemenoff RA. Expression profiling of macrophages reveals multiple populations with distinct biological roles in an immunocompetent Orthotopic model of lung Cancer. J Immunol. 2016;196:2847–59.
Noguera A, Gomez C, Faner R, Cosio B, Gonzalez-Periz A, Claria J, Carvajal A, Agusti A. An investigation of the resolution of inflammation (catabasis) in COPD. Respir Res. 2012;13:101.
Halper-Stromberg E, Yun JH, Parker MM, Tal Singer R, Gaggar A, Silverman EK, Leach S, Bowler RP, Castaldi PJ. Systemic markers of adaptive and innate immunity are associated with COPD severity and Spirometric disease progression. Am J Respir Cell Mol Biol. 2018;58(4):500–9. https://doi.org/10.1165/rcmb.2017-0373OC.
Acknowledgements
Authors thank all patients in the study for their willingness to contribute to medical research and Ms. Gemma Sunyer for her excellent technical support during the study. We also want to thank to technicians (Ms. Mireia Anton and Noemí de Moner) of Flow-cytometry unit in immunology service, for their help and assistance in cytometry. This work was developed at the Centre de Recerca Biomèdica Cellex, Barcelona, Spain.
Declarations
The Ethic Committee of Hospital Clinic de Barcelona approved the study (HCB-2012/7731), and all participants signed their informed consent.
Funding
This work was supported by unrestricted grants from Menarini, Instituto de Salud Carlos III (PI17/000369, PI18/0103), Fondos Feder una nueva manera de hacer Europa. Rosa Faner is recipient of a Miguel Servet Research Program Contract (FEDER, CP16/00039).
Author information
Authors and Affiliations
Contributions
Study conception and design: (RF, GN), data acquisition: (AL-G, TC, LM, SC-R, TG, MJ, MAF), data analysis: (TC, AL-G, RF, GN, MAF, MJ,), manuscript preparation: (RF, TC, AA), manuscript revision: All. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Online Supplement. (DOCX 576 kb)
Additional file 2:
Supplementary Figures. (PDF 1969 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Cruz, T., López-Giraldo, A., Noell, G. et al. Multi-level immune response network in mild-moderate Chronic Obstructive Pulmonary Disease (COPD). Respir Res 20, 152 (2019). https://doi.org/10.1186/s12931-019-1105-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12931-019-1105-z