Abstract
Background
Alpha-1-Antitrypsin (AAT) deficiency (AATD) is a hereditary disorder that manifests primarily as pulmonary emphysema and liver cirrhosis. The clinically most relevant mutation causing AATD is a single nucleotide polymorphism Glu342Lys (Z-mutation). Despite the recommendation to test every COPD patient, the condition remains severely underdiagnosed with a delay of several years between first symptoms and diagnosis. The Grifols’ AlphaKit® QuickScreen is a novel qualitative point-of-care (POC) in vitro screening test developed for the detection of the Z AAT protein in capillary whole blood. The objective of this prospective, international, multi-center, diagnostic, interventional real-world study was to assess the performance of this device for the detection of AATD in test-naïve COPD patients.
Methods
1044 test-naïve COPD patients were recruited from 9 centers in Spain and 10 centers in Germany, ranging from primary to tertiary care. To evaluate the performance of the test, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated compared with the gold standard (genotyping).
Results
Genotyping and phenotyping of all 1019 evaluable samples revealed 4.12% of patients as carriers of at least one Z-allele, while 0.29% carried the homozygous genotype Pi*ZZ. The evaluation of the test’s ability to detect the PiZ protein yielded the following results: specificity 97.8%, sensitivity 73.8%, negative predictive value 98.9%, and positive predictive value 58.5%. All false negatives (n = 11) were heterozygote Pi*MZ samples.
Conclusions
The tested device can be used as an appropriate tool to exclude AATD in primary care and in the overall COPD population, except in patients with a high a-priori- probability of AATD.
Similar content being viewed by others
Background
Alpha-1-Antitrypsin (AAT) deficiency (AATD) is one of the most common hereditary disorders [1]. Clinical manifestations include pulmonary emphysema, liver cirrhosis, c-ANCA positive vasculitis and an inflammatory skin disease called panniculitis [2]. More than 150 genetic variants of coding gene SERPINA1 have been described [3] but the most common deficiency alleles are Pi*Z and Pi*S [4,5,6,7]. In Pi*ZZ, Pi*SZ and some other rare genotypes, the serum level of AAT is deficient [8,9,10].
According to the recent statement by the European Respiratory Society (ERS), diagnostic detection should target individuals with chronic pulmonary disease [11]. Similarly, in 1997 the WHO indicated that all patients with COPD should have their serum level of AAT tested [12]. However, despite these recommendations and the estimation that roughly 1% of subjects with COPD have AATD [2], the condition remains underdiagnosed [13, 14] with a typical delay of several years between first symptoms and diagnosis [15,16,17].
Although a number of detection initiatives and screening programs have been launched over the last several years [18, 19], including in the primary care setting [20], little progress has been made regarding the underdiagnosis and delayed diagnosis of AATD. Limited awareness and knowledge have been identified as the main barriers [21, 22], but a cumbersome testing algorithm may also have a negative impact on the motivation to test for AATD. Currently, AATD testing is based on the combination of different laboratory methods, such as measuring the AAT concentration in serum, followed by a phenotyping and genotyping analysis [23,24,25,26]. As this is a time- and cost-intensive approach, new tools allowing efficient and reliable detection of AATD at the primary care level are warranted.
The AlphaKit® QuickScreen (Diagnostic Grifols, Barcelona, Spain) is a novel qualitative in vitro screening test, with the characteristics of a point-of-care (POC) system, developed for protein Z detection in capillary whole blood [27]. The device is based on the lateral flow assay technology to deliver results within 15–30 min. It uses a mouse monoclonal antibody directed against the Z protein, which are dye labelled. In the event that a Z-variant is present in the blood, the labelled monoclonal antibody would bind to the Z protein in a “sandwich” position, drawing a red line in the reading panel (Z-AAT line). The Z-AAT signal will be positive for all Z allele carriers, regardless if the patient is heterozygous or homozygous for this gene.
In this international, multi-center, diagnostic, interventional real-world study, the performance of this novel device for the detection of AATD in test-naïve COPD patients was assessed.
Methods
Study design and objective
We performed a prospective, multi-center, international, diagnostic real-world study with the objective of assessing the performance of AlphaKit® QuickScreen, a novel vitro diagnostic screening device for AAT deficiency. The performance of the study was monitored by an external clinical research organization (ICON Clinical Research GmbH).
Patients
AATD test-naïve COPD patients were recruited from 9 centers in Spain and 10 centers in Germany, ranging from primary to tertiary care. The samples were processed at the Hospital General Universitari Vall d’Hebron in Barcelona (Spain) and the University Medical Centre Giessen and Marburg (Germany). The inclusion criteria were: i) age ≥ 30; ii) diagnosed with COPD by post- bronchodilator FEV1/FVC < 0.7; iii) being in a stable medical state; and iv)understanding of the study and agreement to give written informed consent. The exclusion criterion was that the participant had not been subjected to any AAT deficiency test before.
Sites identified eligible patients based on their medical records as a pre-screening step. Screening and enrollment occurred on the same day, and only patients who signed the informed consent form were included in the study.
Study device
The AlphaKit® QuickScreen is an immuno-chromatographic, in vitro diagnostic screening test for Z-AAT [27]. Each test requires 30 μL of capillary whole blood. The test is designated for use by qualified healthcare professionals only. The kit contains two variants of a monoclonal antibody against the Z-AAT protein. One is gold-labelled (detector) and the other one is immobilized on the surface on the test strip (capture). The sample (whole blood), together with the running buffer, releases the gold-labelled antibody (detector) out of its matrix. If the examined sample contains the Z-AAT protein, the antibody forms a complex with the analyte (gold-labelled anti-Z-AAT antibody - Z-AAT protein). This complex moves along the test strip by capillary action. At the “Z-AAT”-position, the “capture” anti-Z-AAT antibody is immobilized on the surface and binds the complex specifically via the Z-AAT protein. Depending on the concentration of Z-AAT within the sample, the intensity of the red/purple test line at the “Z-AAT”-position may vary. If the sample does not contain Z-AAT (e.g. Pi*MM), no complex can be formed and therefore no test line will appear. The excess gold-labelled antibodies bind at the control line (“Control”) indicating that the chromatography worked properly.
For Z-AAT detection, 30 μL of finger stick capillary blood were added into the sample reservoir of the screening device and, after waiting 1–2 min, 3 drops of running buffer were applied. Within 15–30 min, the results were read in the display.
Testing device performance evaluation
Test performance was evaluated by analyzing the samples with the testing device and comparing the results with those obtained by genotyping.
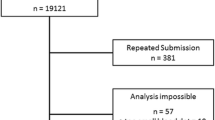
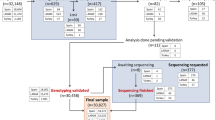
Both positive and negative test results were confirmed independently and in a blinded manner by the central laboratory. For genotyping, the AlphaKit® (Diagnostic Grifols, Barcelona, Spain) test was used to collect finger stick capillary blood samples on the filter paper. After allowing the blood sample to dry for 1 h, the specimens were mailed to the testing laboratories in Marburg (Germany) or Barcelona (Spain) where genotyping for S and Z by means of the PCR technology and AAT level measurements were performed. The exact phenotype was determined in samples where the S or Z mutation was present (according to genotyping) or when the DBS-derived AAT level was < 110 mg/dl [23, 28]. The testing algorithm is summarized in Fig. 1.
The testing algorithm is summarized. In both countries (Spain and Germany), the initial analysis consisted of the screening device (AlphaKit® QuickScreen) and the acquisition of blood on DBS (dried blood spot – AlphaKit). If the DBS-derived AAT serum level was ≥110 mg/dl and genotyping for the Z- and S-allele was negative, AATD was considered as excluded. All other samples underwent phenotyping to obtain a final result. DBS: dried blood spot; WBS: whole blood sample
Data analysis
The sample size calculation was based on the assumptions that i) the percentage of COPD subjects with severe AATD Pi*ZZ was likely to be less than 1% in an unselected population; ii) the percentage of COPD subjects with heterozygous AATD Pi*MZ was assumed to be between 2 and 5%; iii) the percentage of COPD subjects with heterozygous AATD Pi*SZ was likely to be less than 1%, and iii) the total percentage of positive (any genotype combination with a Z-protein) with the tested screening device was expected to be between 2 and 5%. Therefore the number of subjects planned to be tested was 1000 (500 per country) with the expectation that 50 subjects would carry the Z-allele. The total number of enrolled subjects could be adjusted to exceed 1000, until 50 Z-positive samples were detected.
We performed descriptive statistics displaying continuous variables as mean ± standard deviation (SD), and categorical variables by number and percentage. To evaluate the performance of the test, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated compared with the gold standard (genotyping). Discrepancies were shown as numbers of false positive (FP) and false negative (FN) results. Causes of discrepancies were analyzed.
From the study-derived sensitivity and specificity, a variety of predictive values were calculated to simulate the test-performance under a number of clinical scenarios with different prevalence (pre-test probability).
SPSS Version 22 (IBM, Ehningen, Germany), and GraphPad Prism Version 7 (GraphPad Software, Inc., La Jolla, USA) were used for calculations.
Results
Patients characteristics
One thousand and forty four test-naïve COPD patients that met the inclusion criteria were enrolled. Twenty-five of them were excluded: 20 due to missing results from the tested screening device or genotyping and 5 because the screening test was performed incorrectly.
The full analysis set consisted of 1019 patients, 549 from Germany and 470 from Spain. The mean age was 66.3 ± 9.7 years, the mean FEV1 was 53.0 ± 20.3%predicted, and the majority of the test population was male (69.3%). Baseline demographic and clinical characteristics of patients by country are shown in Table 1.
Prevalence of pi*Z positive samples in the study population
Genotyping and phenotyping of all 1019 samples indicated that 4.12% of patients carried at least one Z-allele. The percentage of patients who carried the homozygous genotype Pi*ZZ was 0.29%. Results were similar in patients from Germany and Spain. Details for the S- and Z-allele by country are shown in Table 2.
Testing device performance
Results of test performance for detecting Z protein showed high specificity and NPV of the tested screening device when compared to genotyping (97.8 and 98.9%, respectively), while sensitivity and PPV resulted lower (73.8 and 58.5%, respectively). Detailed results of truly positive, truly negative, falsely positive and falsely negative samples are shown in Table 3. The analysis of false negatives showed that all 11 patients were heterozygote Pi*MZ.
As the predictive values are based on the prevalence of the disease in the test population, we simulated the test-performance under a number of clinical scenarios (reported in the literature) with different prevalence (pre-test probability). The negative predictive value was high (> 98%) in unselected COPD populations (prevalence values taken from the literature) as well as in our study population, and decreased to lower numbers in a population with a very high pre-test probability (Table 4).
Discussion
In this real-world study, we assessed the performance of a novel POC technology-based tool for the detection of AATD in test-naïve COPD patients. The tested device exhibited a high specificity and a moderate sensitivity.
More than 1000 samples were obtained from test-naïve COPD patients and compared to the gold standard, a combination of serum level, genotyping and – where applicable – phenotyping. The intended sample size was met and exceeded. The demographic and clinical profile of the studied population was the expected for the underlying pathology. Similarities between patients from both participating countries indicated that likely no selection bias took place.
The prevalence of samples carrying at least one Z-mutation was 4.1% in the overall test population (4.7% in Germany and 3.4% in Spain). The results of other targeted screening studies vary widely [29], but recent screening studies in Germany and Spain in test-naïve populations similar to our populations yielded remarkably similar results. In a study on 971 COPD patients in Spain, a total of 2.9% carriers of a Z allele was found [30], and in a population-based study, using health-care records in Catalonia, Barrecheguren et al. found that 5.3% of the test population exhibited an intermediated AAT serum level (50–100 mg/dl), most likely due to the presence of one Z-allele [31]. Similarly, Wencker et al. found a prevalence of 3.7% Pi*MZ in a German targeted test population [32].
In our study, the tested device performed with a specificity of 97.8% and sensitivity of 73.8%. In the prevalence observed, this corresponded to a positive predictive value of 58.5% and a negative predictive value of 98.9%. The positive predictive value of 58,5% clearly indicates that further testing (genotyping/phenotyping) is absolutely mandatory to confirm and clarify a positive test result. However, for a screening test, the most important indicator of performance is the negative predictive value, as it describes the level of certainty with which the disease can be excluded after having obtained a negative test result. Comparing the examined screening device with other tests that are widely used in clinical practice, it is interesting to note that for example procalcitonin (in combination with CRB-65) exhibited a negative predictive value of 98.8%, also [33]. Furthermore, because all false negatives were heterozygote Pi*MZ, all Pi*ZZ and Pi*SZ have been correctly identified.
The intermediate sensitivity of 73.8% may limit the applicability of the test in populations where the a-priory-probability is higher than in our current study population. As illustrated in Table 4, the test sufficiently rules out AATD in our test population as well as in other unselected COPD populations where the prevalence of the Z-allele may be around 5% [34], which would result in a negative predictive value of 98.5%. However, in clinical scenarios with an a-priory-probability of greater than 16.4% (examples could include COPD patients with an index case as a first degree relative or a young never-smoker with high-grade basal emphysema) the false negative rate would increase to values > 5%, probably judged as insufficient for use in that specific patient population. If a false negative rate of 10% would be judged as still acceptable, the prevalence of the population in which the test would be applicable, could be as high as 29.3%.
A further test-immanent limitation is that only deficient genotypes containing at least one Z allele can be detected [27]. In general, combinations of deficiency alleles without a Z-mutation such as Null/Null or S/Null are extremely rare [9, 35]. With the exception of countries like Italy where the prevalence of non-Z-mutations is considerably higher [36, 37], this may not limit the usability of the test.
A combination of different laboratory methods has been described as the optimal strategy for the diagnosis of AATD [23,24,25,26], and quantitative measurements of the AAT serum level are used widely as the initial screening test [38, 39], but the determination requires considerable time. Furthermore, as an acute phase reactant, levels of AAT may vary depending on clinical conditions. The tested screening device should not be seen as a diagnostic tool but could be used as a screening tool. Phenotyping and genotyping are necessary upon indication of a positive result. The test kit may have considerable potential to increase the overall AATD detection rate, simplifying the screening pathway. Early diagnosis is essential, as some individuals will qualify for augmentation therapy with AAT as well as benefit from lifestyle modifications.
Overall, these results demonstrate the tested device to be useful in primary care and in the overall COPD population, except in patients with a high a-priori-probability of AATD. In these patients, determination of the serum level and further diagnostic steps would be required.
Conclusions
We performed an international, multi-center, diagnostic, interventional real-world study to assess the performance of AlphaKit® QuickScreen, a novel device for the screening of AATD in test-naïve COPD patients. With a negative predictive value of 98.9%, we conclude this novel screening device to be useful in primary care and in the overall COPD population, except in patients with a markedly increased a-priori-probability of AATD.
References
Stoller JK, Aboussouan LS. Alpha1-antitrypsin deficiency. Lancet. 2005;365:2225–36.
Silverman EK, Sandhaus RA. Clinical practice. Alpha1-antitrypsin deficiency. N Engl J Med. 2009;360:2749–57.
Demeo DL, Silverman EK. Alpha1-antitrypsin deficiency. 2: genetic aspects of alpha(1)-antitrypsin deficiency: phenotypes and genetic modifiers of emphysema risk. Thorax. 2004;59:259–64.
Blanco I, de Serres FJ, Fernandez-Bustillo E, Lara B, Miravitlles M. Estimated numbers and prevalence of PI*S and PI*Z alleles of alpha1-antitrypsin deficiency in European countries. Eur Respir J. 2006;27:77–84.
Greene CM, Miller SD, Carroll T, McLean C, O'Mahony M, Lawless MW, O'Neill SJ, Taggart CC, McElvaney NG. Alpha-1 antitrypsin deficiency: a conformational disease associated with lung and liver manifestations. J Inherit Metab Dis. 2008;31:21–34.
Lomas DA, Evans DL, Finch JT, Carrell RW. The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature. 1992;357:605–7.
Fregonese L, Stolk J. Hereditary alpha-1-antitrypsin deficiency and its clinical consequences. Orphanet J Rare Dis. 2008;3:16.
de Serres FJ, Blanco I, Fernandez-Bustillo E. Estimating the risk for alpha-1 antitrypsin deficiency among COPD patients: evidence supporting targeted screening. COPD. 2006;3:133–9.
Blanco I, Bueno P, Diego I, Perez-Holanda S, Casas-Maldonado F, Esquinas C, Miravitlles M. Alpha-1 antitrypsin pi*Z gene frequency and pi*ZZ genotype numbers worldwide: an update. Int J Chron Obstruct Pulmon Dis. 2017;12:561–9.
Blanco I, Bueno P, Diego I, Perez-Holanda S, Lara B, Casas-Maldonado F, Esquinas C, Miravitlles M. Alpha-1 antitrypsin pi*SZ genotype: estimated prevalence and number of SZ subjects worldwide. Int J Chron Obstruct Pulmon Dis. 2017;12:1683–94.
Miravitlles M, Dirksen A, Ferrarotti I, Koblizek V, Lange P, Mahadeva R, McElvaney NG, Parr D, Piitulainen E, Roche N, et al. European Respiratory Society statement: diagnosis and treatment of pulmonary disease in alpha1-antitrypsin deficiency. Eur Respir J. 2017;50
WHO. Alpha 1-antitrypsin deficiency: memorandum from a WHO meeting. Bull World Health Organ. 1997;75:397–415.
de Serres FJ, Blanco I. Prevalence of alpha1-antitrypsin deficiency alleles PI*S and PI*Z worldwide and effective screening for each of the five phenotypic classes PI*MS, PI*MZ, PI*SS, PI*SZ, and PI*ZZ: a comprehensive review. Ther Adv Respir Dis. 2012;6:277–95.
Stoller JK, Brantly M. The challenge of detecting alpha-1 antitrypsin deficiency. COPD. 2013;10(Suppl 1):26–34.
Campos MA, Wanner A, Zhang G, Sandhaus RA. Trends in the diagnosis of symptomatic patients with alpha1-antitrypsin deficiency between 1968 and 2003. Chest. 2005;128:1179–86.
Kohnlein T, Janciauskiene S, Welte T. Diagnostic delay and clinical modifiers in alpha-1 antitrypsin deficiency. Ther Adv Respir Dis. 2010;4:279–87.
Stoller JK, Sandhaus RA, Turino G, Dickson R, Rodgers K, Strange C. Delay in diagnosis of alpha1-antitrypsin deficiency: a continuing problem. Chest. 2005;128:1989–94.
Chorostowska-Wynimko J. Targeted screening programmes in COPD: how to identify individuals with alpha1-antitrypsin deficiency. Eur Respir Rev. 2015;24:40–5.
Corda L, Bertella E, Pini L, Pezzini A, Medicina D, Boni E, Guerini M, Trivella S, Grassi V, Tantucci C. Diagnostic flow chart for targeted detection of alpha1-antitrypsin deficiency. Respir Med. 2006;100:463–70.
Molina J, Flor X, Garcia R, Timiraos R, Tirado-Conde G, Miravitlles M. The IDDEA project: a strategy for the detection of alpha-1 antitrypsin deficiency in COPD patients in the primary care setting. Ther Adv Respir Dis. 2011;5:237–43.
Esquinas C, Barrecheguren M, Sucena M, Rodriguez R, Fernandez S, Miravitlles M. Practice and knowledge about diagnosis and treatment of alpha-1 antitrypsin deficiency in Spain and Portugal. BMC Pulm Med. 2016;16
Greulich T, Ottaviani S, Bals R, Lepper PM, Vogelmeier C, Luisetti M, Ferrarotti I. Alpha1-antitrypsin deficiency - diagnostic testing and disease awareness in Germany and Italy. Respir Med. 2013;107:1400–8.
Bals R, Koczulla R, Kotke V, Andress J, Blackert K, Vogelmeier C. Identification of individuals with alpha-1-antitrypsin deficiency by a targeted screening program. Respir Med. 2007;101:1708–14.
Ferrarotti I, Scabini R, Campo I, Ottaviani S, Zorzetto M, Gorrini M, Luisetti M. Laboratory diagnosis of alpha1-antitrypsin deficiency. Transl Res. 2007;150:267–74.
Snyder MR, Katzmann JA, Butz ML, Wiley C, Yang P, Dawson DB, Halling KC, Highsmith WE, Thibodeau SN. Diagnosis of alpha-1-antitrypsin deficiency: an algorithm of quantification, genotyping, and phenotyping. Clin Chem. 2006;52:2236–42.
Miravitlles M, Herr C, Ferrarotti I, Jardi R, Rodriguez-Frias F, Luisetti M, Bals R. Laboratory testing of individuals with severe alpha1-antitrypsin deficiency in three European centres. Eur Respir J. 2010;35:960–8.
Vogelmeier C, Soriano J, Janciauskiene S, Crystal RG, Ferrarotti I, Carroll T. Alpha-1-antitrypsin deficiency: honouring the past and embracing the future. Grifols' symposium report. Expert Rev Respir Med. 2013;(Special Edition):1–10.
Rodriguez F, Jardi R, Costa X, Cotrina M, Galimany R, Vidal R, Miravitlles M. Rapid screening for alpha1-antitrypsin deficiency in patients with chronic obstructive pulmonary disease using dried blood specimens. Am J Respir Crit Care Med. 2002;166:814–7.
Greulich T, Vogelmeier CF. Alpha-1-antitrypsin deficiency: increasing awareness and improving diagnosis. Ther Adv Respir Dis. 2016;10:72–84.
de la Roza C, Rodriguez-Frias F, Lara B, Vidal R, Jardi R, Miravitlles M. Results of a case-detection programme for alpha1-antitrypsin deficiency in COPD patients. Eur Respir J. 2005;26:616–22.
Barrecheguren M, Monteagudo M, Simonet P, Llor C, Rodriguez E, Ferrer J, Esquinas C, Miravitlles M. Diagnosis of alpha-1 antitrypsin deficiency: a population-based study. Int J Chron Obstruct Pulmon Dis. 2016;11:999–1004.
Wencker M, Marx A, Konietzko N, Schaefer B, Campbell EJ. Screening for alpha1-pi deficiency in patients with lung diseases. Eur Respir J. 2002;20:319–24.
Kruger S, Ewig S, Marre R, Papassotiriou J, Richter K, von Baum H, Suttorp N, Welte T, Group CS. Procalcitonin predicts patients at low risk of death from community-acquired pneumonia across all CRB-65 classes. Eur Respir J. 2008;31:349–55.
Rahaghi FF, Sandhaus RA, Brantly ML, Rouhani F, Campos MA, Strange C, Hogarth DK, Eden E, Stocks JM, Krowka MJ, Stoller JK. The prevalence of alpha-1 antitrypsin deficiency among patients found to have airflow obstruction. COPD. 2012;9:352–8.
Greulich T, Nell C, Herr C, Vogelmeier C, Kotke V, Wiedmann S, Wencker M, Bals R, Koczulla AR. Results from a large targeted screening program for alpha-1-antitrypsin deficiency: 2003 - 2015. Orphanet J Rare Dis. 2016;11:75.
Ferrarotti I, Baccheschi J, Zorzetto M, Tinelli C, Corda L, Balbi B, Campo I, Pozzi E, Faa G, Coni P, et al. Prevalence and phenotype of subjects carrying rare variants in the Italian registry for alpha1-antitrypsin deficiency. J Med Genet. 2005;42:282–7.
Ferrarotti I, Carroll TP, Ottaviani S, Fra AM, Brien O, Molloy K, Corda L, Medicina D, Curran DR, McElvaney NG, Luisetti M. Identification and characterisation of eight novel SERPINA1 null mutations. Orphanet J Rare Dis. 2014;9:172.
Steiner SJ, Gupta SK, Croffie JM, Fitzgerald JF. Serum levels of alpha1-antitrypsin predict phenotypic expression of the alpha1-antitrypsin gene. Dig Dis Sci. 2003;48:1793–6.
Bornhorst JA, Greene DN, Ashwood ER, Grenache DG. alpha1-antitrypsin phenotypes and associated serum protein concentrations in a large clinical population. Chest. 2013;143:1000–8.
Acknowledgements
We thank the principal investigators of all participating centers: Adolfo Baloira, Robert Bals, Miriam Barrecheguren, Salvator Diaz-Lobato, Cristina Esquinas, Cristobal Esteban, Felix Herth, Clemens Kelbel, Klaus Kenn, María Teresa Martinez, Augustin Medina, Carmen Montero, Conception Morales, Ulrich Steinhauser, Hartmut Timmermann, Augustin Valido, Walter Vorderstraße, Tobias Welte, Andreas Wilke. Bettina Dreger PhD, Carme Romera PhD, and Jordi Bozzo PhD CMPP (Grifols) are acknowledged for their assistance in the preparation of the manuscript, under the direction of the authors. Marion Trautmann (Grifols) was involved in the preparation of the study protocol and execution of the trial. ICON Clinical Research GmbH provided monitoring, data management, statistical input and programming, analyses as well as medical writing support.
With great sadness, we commemorate Silvia Gimeno-Pustoslemsek who co-designed the study and was one of the driving forces during the conduct. On the 23rd of November 2017, she passed away. We miss her.
Funding
The study was sponsored by Grifols (Colmarer Straße 22, Frankfurt, Germany), manufacturer of AlphaKit® Quickscreen.
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available due to more secondary analyses but are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
TG, FR-F, CFV, and MM designed the study. TG, FR-F, IB, and AK carried out the laboratory analyses. TG and MM wrote the manuscript in consultation with CFV. All authors provided critical feedback and helped significantly to shape the analysis and manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the ethics committee at the University of Marburg for all centers in Germany, and by the ethics committee at the University Hospital Vall d’Hebron in Barcelona for all centers in Spain.
Only patients who signed the informed consent form were included in the study.
Consent for publication
Not applicable.
Competing interests
The study was sponsored by Grifols (Colmarer Straße 22, Frankfurt, Germany), manufacturer of AlphaKit® Quickscreen.
TG reports personal fees from Astra Zeneca, Berlin-Chemie, Boehringer Ingelheim, Chiesi, CSL-Behring, GlaxoSmithKline, Novartis. He reports grants and personal fees from Grifols.
FR-F reports no conflicts of interest.
IB reports no conflicts of interest.
AK reports no conflicts of interest.
CFV reports grants and personal fees from AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Grifols, Novartis. He reports personal fees from CSL Behring, Chiesi, Menarini, Mundipharma, Teva, Cipla.
MM has received speaker fees from Boehringer Ingelheim, Chiesi, Cipla, Menarini, Rovi, Grifols and Novartis, and consulting fees from Boehringer Ingelheim, Chiesi, GlaxoSmithKline, Gebro Pharma, CSL Behring, Novartis and Grifols.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Greulich, T., Rodríguez-Frias, F., Belmonte, I. et al. Real world evaluation of a novel lateral flow assay (AlphaKit® QuickScreen) for the detection of alpha-1-antitrypsin deficiency. Respir Res 19, 151 (2018). https://doi.org/10.1186/s12931-018-0826-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12931-018-0826-8