Abstract
Background
Cancer susceptibility germline mutations are associated with pancreatic ductal adenocarcinoma (PDAC). However, the hereditary status of PDAC and its impact on survival is largely unknown in the Asian population.
Methods
Exome sequencing was performed on 527 blood samples from PDAC individuals and analyzed for mutations in 80 oncogenic genes. Pathogenic and likely pathogenic (P/LP) germline variants were diagnosed according to the ACMG variant classification categories. The association between germline homologous recombination gene mutations (gHRmut, including BAP1, BRCA1, BRCA2, PALB2, ATM, BLM, BRIP1, CHEK2, NBN, MUTYH, FANCA and FANCC) and the treatment outcomes was explored in patients with stage III/IV diseases treated with first-line (1L) platinum-based versus platinum-free chemotherapy.
Results
Overall, 104 of 527 (19.7%) patients carried germline P/LP variants. The most common mutated genes were BRCA2 (3.60%), followed by ATR (2.66%) and ATM (1.9%). After a median follow-up duration of 38.3-months (95% confidence interval, 95% CI 35.0–43.7), the median overall survival (OS) was not significantly different among patients with gHRmut, non-HR germline mutations, or no mutation (P = 0.43). Among the 320 patients with stage III/IV disease who received 1L combination chemotherapy, 32 (10%) had gHRmut. Of them, patients receiving 1L platinum-based chemotherapy exhibited a significantly longer median OS compared to those with platinum-free chemotherapy, 26.1 months (95% CI 12.7–33.7) versus 9.6 months (95% CI 5.9–17.6), P = 0.001. However, the median OS of patients without gHRmut was 14.5 months (95% CI 13.2–16.9) and 12.6 months (95% CI 10.8–14.7) for patients receiving 1L platinum-based and platinum-free chemotherapy, respectively (P = 0.22). These results were consistent after adjusting for potential confounding factors including age, tumor stage, performance status, and baseline CA 19.9 in the multivariate Cox regression analysis.
Conclusions
Our study showed that nearly 20% of Taiwanese PDAC patients carried germline P/LP variants. The longer survival observed in gHRmut patients treated with 1L platinum-based chemotherapy highlights the importance of germline testing for all patients with advanced PDAC at diagnosis.
Similar content being viewed by others
Background
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive disease with an increasing incidence and mortality globally. To date, PDAC is the third leading cause of cancer mortality in the United States with a five-year survival rate of less than 12% [1]. The poor prognosis of PDAC is mainly due to delayed diagnosis, a lack of feasible detection markers, low resectability, a high recurrence rate, high resistance to chemotherapy and radiotherapy, and limited available effective therapeutic agents [2]. The mortality of major cancer types including lung cancer, breast cancer and colorectal cancer, is steadily decreasing after the introduction of novel target therapy and immunotherapy [3]. Immune checkpoint inhibitors have demonstrated promising efficacy in a variety of cancer types, but it adds limited clinical benefits when combining with standard chemotherapy in PDAC [4]. Currently, there are no effective target therapies in PDAC except poly (ADP-ribose) polymerase (PARP) inhibitors in a small subset of metastatic PDAC patients with germline BRCA1 or BRCA2 mutations [5]. As a result, PDAC is predicted to become the second leading cause of cancer death in the United States by 2026 and in Germany by 2030 [3, 6]. A screening program for early diagnosis, new treatment strategies, and the discovery of target-specific drugs to decrease deaths from PDAC are urgently needed.
Given that most PDAC patients are diagnosed at advanced stage, screening programs for all comers may not cost-effective due to the relatively low incidence of PDAC. Identification of high-risk individuals is a more reasonable approach for screening to detect early stage PDAC and yield better treatment outcomes. Individuals with a family history of cancer were shown to have a greatly elevated risk of developing cancer, especially so-called BRCA-associated hereditary cancers, including breast, ovary, prostate, and pancreatic cancer [7,8,9]. Notably, approximately 5–10% of PDAC patients in the United States and Japan have a family history of the disease [10, 11]. The alterations in cancer susceptible genes such as BRCA1, BRCA2, PALB2, STK11, PRSS1, and ATM have been linked to increased lifetime risks of PDAC [12,13,14]. Family members of PDAC patients with germline P/LP mutations in hereditary cancer susceptible genes are the candidates for active screening programs, such as Pancreatic Cancer Early Detection (PRECEDE) Consortium [15].
The development of a new drug usually requires enormous amounts of time and money. Through the concept of precision medicine, using the right drug for the right patient at the right time, the outcomes of PDAC can be more efficiently improved, as demonstrated in the Know Your Tumor program [16]. Germline mutations of BRCA1, BRCA2, or PALB2 were associated with improved responses to and survival under platinum-based chemotherapy [17, 18]. There is increasing evidence that PDAC patients with mutations in other homologous recombination (HR) genes can also benefit from platinum-based chemotherapy [19, 20]. Identifying patients with germline mutations in cancer susceptibility genes, especially HR genes, not only assist in screening high-risk familiar members but also benefit proper treatment selection.
Current genomic studies have investigated the germline mutation landscape of PDAC patients based on whole genome sequencing and exome sequencing. However, the hereditary mutation genes in the Asian population are poorly described. In this study, we performed exome sequencing on 527 Taiwanese PDAC blood samples to identify possible germline mutations in cancer susceptibility genes and to explore the association between germline HR genes mutations and outcomes of advanced PDAC receiving first-line (1L) platinum-based versus platinum-free chemotherapy.
Materials and methods
Sample acquisition and questionnaires
A multicenter prospective observation cohort that recruited PDAC patients from the National Cheng Kung University Hospital and the Kaohsiung Medical University Hospital was established in 2013 [21]. Patient recruitment is still ongoing. The current analysis included 527 patients who consented between 2013 and 2021. The review process of clinical information was retrospectively registered on January 26, 2023 (ClinicalTrials.gov Identifier: NCT05700188). Patients with multiple cancer types were excluded from this study. All patients provided written informed consent, donated 20 ml blood samples, and answered a detailed questionnaire about the age at diagnosis of any cancer and the types of cancer diagnosed in their first- and second-degree relatives. Questionnaires were completed by patients at home and sent back to the physicians.
DNA extraction, exome sequencing, and data processing
Genomic DNA was extracted from peripheral blood samples using a Wizard® Genomic DNA Purification Kit (Promega). All samples were processed using two different WES enrichment platforms: Nextera Flex for Enrichment, an Exome panel (Illumina), and an Accel-NGSR 2S Hyb DNA Library Kit (Swift). In total, 50–1000 ng of DNA was sheared into short fragments using enzymatically (Illumina) and Covaris M220 Focused-ultrasonicator (Swift). Size selection, sequence capturing, enrichment, and elution were performed according to the manufacturer’s instructions. The size distribution of DNA libraries was then measured using 4150 TapeStation (Agilent). Finally, the validated DNA libraries were sequenced on an Illumina sequencing platform (NovaSeq 6000) with 150 bp paired-end reads. The read pair data (fastq) of each sample were generated from the sequencing system. The resulting reads were then aligned to the reference human genome sequence (GRCh37/hg19), and nucleotide variant calling was performed using the DRAGEN Enrichment app (ver 3.6.3) in the Illumina basespace sequence hub. The VCF files from the DRAGEN Enrichment app were annotated and analyzed in the CLC Genomics Workbench (ver 20) software with default parameters.
Data analysis and variant characterization
Eighty oncogenic candidate genes associated with germline mutations were chosen for this study based on a literature review and COSMIC cancer gene census. Selected genes were divided into four nonoverlapping categories according to previous reports. Mutations in 12 genes, including BAPI, BRCA1, BRCA2, PALB2, ATM, BLM, BRIP1, CHEK2, NBN, MUTYH, FANCA, and FANCC, were considered as homologous recombination (HR) genes (Additional file 1: Table S1) [22,23,24,25,26,27,28,29,30,31,32,33]. Variants were retained if they passed the quality criteria (variant count of ≥ 2 and variant allele frequency between 20 and 80%). Pathogenic probabilities according to The American College of Medical Genetics and Genomics (ACMG) variant classification categories were determined by the VarSome (ver.11.4) prediction database. Only pathogenic, likely pathogenic, and variants of uncertain significance (VUS) were analyzed in our study. Pathogenic and likely pathogenic variants were also filtered if the minor allele frequency (MAF) was > 1% in the 1000 Genomes database, Genome Aggregation Database (gnomAD), and dbSNP database. Annotated variants were further assessed in common databases such as ClinVar and HGMD.
Statistical analysis
Fisher’s exact test was performed to determine the cancer types among those with family cancer history between carriers and non-carriers of pathogenic variants. A paired t-test was applied to compare the ages of patients with or without family history of BRCA-associated hereditary cancers, with or without germline mutation and with or without gHRmut. Overall survival was calculated from diagnosis to death due to any cause or censored in the case of loss to follow-up or data cut-off. Two-sided P < 0.05 was considered statistically significant.
Results
Population characteristics
The clinical features of 527 PDAC patients are summarized in Table 1. In total, 298 males (56.5%) and 229 females (43.5%) participated in this study. The median age of diagnosis of the entire cohort was 62.5 years, ranging from 27.0 to 93.9 years. A total of 48 patients (9.1%) had stage I disease, 111 (21.1%) had stage II disease, 120 (22.8%) had stage III disease, and 248 (47.1%) had stage IV disease upon diagnosis. The most common primary location of the tumor was in the head (37.0%), followed by the body (20.5%), tail (17.8%), and uncinate process (3%). The remaining 114 (21.6%) cases had tumors located in overlapping areas. Family history of any cancer, which included first- and second-degree relatives, was observed in 54.3% of patients.
Germline mutational landscape of pancreatic cancer
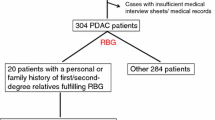
To investigate the susceptibility of germline mutational genes in pancreatic cancer, we performed exome sequencing of all PDAC patients in the study cohort. The mean coverage in the exome sequencing data was 62 (range, 19–164). In total, 80 genes were selected because of the potential effects of germline mutations, and the predicted pathogenicity of variants was determined according to ACMG variant classification categories. As shown in Fig. 1, a total of 20,712 germline variants were identified, and 12,625 variants passed the quality control. Of these, 62 (0.49%) were considered pathogenic variants, 58 (0.46%) were likely pathogenic variants, 410 (3.25%) were variants of uncertain significance, 744 (5.89%) were likely benign, and 11,351 (89.91%) were benign variants. Among 527 PDAC patients, a total number of 104 (19.7%) patients carried 120 P/LP variants, 15 patients had two or more P/LP variants. In our classification categories, 47 (39.2%) P/LP variants were considered to have high penetrance, followed by 40 (33.3%) with moderate penetrance, 22 (18.3%) with low penetrance, and 11 (9.2%) indicating a recessive condition (Fig. 2A). A total number of 120 P/LP variants included 21 (17.5%) missense mutations, 13 (10.8%) nonsense mutations, 79 (65.8%) frameshift mutations, 5 (4.2%) inframe mutations, and 2 (1.7%) start codon mutations (Fig. 2B). The most common mutated genes were BRCA2 (3.6%), followed by ATR (2.66%) and ATM (1.9%) (Fig. 2C). Here, we summarize the most frequent P/LP variants (case ≥ 3) in Table 2, which includes ATM (p.Lys468fs), ATR (p.Ile774fs), and WRN (p.Lys168fs). Detailed characteristics of the 120 pathogenic and likely pathogenic mutations carried in 104 patients were summarized in Additional file 1: Table S2.
The distribution of 120 pathogenic and likely pathogenic (P/LP) germline mutations in PDAC patients. A The frequency of P/LP mutation genes in four categories (low-, moderate-, high- and recessive penetrance). B The frequency of the mutational type of P/LP mutation genes. C The landscape of P/LP mutation genes in patients. The distribution of mutational type was included in the figure
Associations between family cancer history and age distribution in PDAC patients
Overall, 286 of 527 (54.3%) patients had a family cancer history in our study (Table 1) and the most common types of cancer among the families were liver cancer (13.7%), lung cancer (9.1%), pancreatic cancer (8.4%), colon cancer (7.8%), and breast cancer (5.9%). A family history of pancreatic cancer was identified in 8.7% (9/104) of P/LP variants carriers and 8.3% (35/423) of non-carriers. Detailed family history information was shown in Additional file 1: Table S3. Excluding five patients with unknown family history, 82 of 522 (15.7%) patients had a family history of BRCA-associated hereditary cancers including breast, ovary, prostate, and pancreatic cancer. The diagnosed ages of patients with a family history of these four cancer types were significantly younger than the ages of those without a family history (median age 59.6 vs. 63.0 years old, P = 0.0094) (Fig. 3A). Conversely, the diagnosed ages of patients with germline mutations in the 80 oncogenic candidate genes or gHRmut were not significantly different than those of their counterparts (Fig. 3B, C).
Distribution of pancreatic cancer diagnosed age. A Patients with or without family history of breast, ovary, prostate, and pancreatic cancer. B Patients with or without germline mutation in 80 oncogenic candidate genes. C Patients with or without germline mutation in 12 homologous recombination genes
Impact of germline HR gene alterations on chemotherapy efficacy
As of December 31, 2021, the median duration of follow-up was 38.3 months (95% confidence interval [95% CI] 35.0–43.7). P/LP variant carriers tend to have a better median OS than non-carriers in both stage I/II (33.2 months vs. 25.1 months, P = 0.9) and stage III/IV (18.0 months vs. 13.2 months, P = 0.062) cancer (Additional file 1: Fig. S1). Patients with gHRmut exhibited a numerically better median OS of 26.1 months (95% CI 13.7–32.3), compared to 18.7 months (95% CI 12.4–25.2) in those with non-HR germline mutations and 15.9 months (95% CI 14.1–18.6) in patients without germline mutations, P = 0.43 (Fig. 4A).
Outcome of PDAC patient in homologous recombination (HR) gene mutations status. A Median overall survival of 527 PDAC patients with germline HR gene mutations, other gene mutations or no mutation. B Median overall survival of 320 stage III/IV PDAC patients with or without HR gene mutations treated with 1L platinum-based or platinum-free chemotherapy
To further investigate the correlation between germline mutations and chemotherapy efficacy, patients who received first-line chemotherapy were identified for detailed analysis. A total of 159 patients with stage I/II PDAC were excluded, as chemotherapy is not the standard of care at this stage. Among the 368 patients with stage III/IV diseases, 3 patients who received unknown treatments at other hospital, 2 untreated patients, and 43 fragile patients who underwent non-standard including gemcitabine monotherapy were excluded. Finally, 320 patients with stage III/IV diseases underwent first-line combination chemotherapy, including 32 patients (10%) with gHRmut and 288 patients with germline HR wild-type (gHRwt), were included in the analysis (Figs. 1 and 4B). The baseline characteristics, including sex, age, tumor location, stage, family history, ECOG, albumin, CA-19.9 and 1L chemotherapy regimens, platinum-based or platinum-free chemotherapy were not significantly different between patients with or without gHRmut (Table 3). Overall, 214 patients were treated with 1L platinum-based chemotherapy, SLOG (S1, leucovorin, oxaliplatin and gemcitabine) in 153 (47.8%), modified FOLFIRINOX in 34 (10.6%) and GOFL (gemcitabine, oxaliplatin, fluorouracil, and leucovorin) in 27 (8.4%). A total of 106 patients received 1L platinum-free chemotherapy, including gemcitabine plus nab-paclitaxel in 54 (16.9%) and gemcitabine plus S1 in 52 (16.3%) (Table 3).
The median OS of patients with gHRmut receiving 1L platinum-based chemotherapy was significantly better than those receiving platinum-free chemotherapy, 26.1 months (95% CI 12.7—33.7) versus 9.6 months (95% CI 5.9–17.6) (P = 0.001, Fig. 4B). However, the median OS was not significantly different between gHRwt patients treated with 1L platinum-based and platinum-free chemotherapy, with a median OS of 14.5 months (95% CI 13.2–16.9) and 12.6 months (95% CI 10.8–14.7), respectively (P = 0.22, Fig. 4B). All baseline characteristics were not significantly different for those with gHRmut receiving 1L platinum-based or platinum-free chemotherapy (Additional file 1: Table S4).
Overall, 37 of 107 (34.6%) patients with stage III disease underwent conversion surgery after first-line chemotherapy, including 6 of 13 patients (46.2%) in the gHRmut group and 31 of 94 patients (33.0%) in the gHRwt group (Fisher’s exact test, P = 0.37). The median OS of patients with and without conversion surgery was 33.7 months (95% CI 29.7–NE) and 16.5 months (95% CI 13.7–20.2), respectively. (Additional file 1: Fig. S2).
Adjustment of confounding factors
Potential confounding factors were adjusted using a multivariable Cox regression analysis, which included age, gender, tumor location, stage, ECOG PS, baseline albumin, baseline CA 19.9, year of diagnosis, and enrollment in the clinical trial. After this adjustment, age, ECOG PS, and baseline CA 19.9 were identified as significant prognostic factors. Additionally, patients with gHRmut treated with 1L platinum-based chemotherapy continued to show a significant improvement in survival compared to those treated with platinum-free chemotherapy, with a hazard ratio of 0.41 (95% CI 0.18–0.94, P = 0.036) (Fig. 5).
Due to the delayed reimbursement of nab-paclitaxel (reimbursed in 2019) and FOLFIRINOX (reimbursed in 2021), a great proportion of patients in the current study participated in investigator-initiated trials (IITs), as the treatment options were very limited in Taiwan at that time (Additional file 1: Fig. S3) [34,35,36,37,38,39,40,41,42]. Therefore, during the study period (2013–2020), the majority of patients received SLOG, either in IITs or in daily practice, while nab-paclitaxel plus gemcitabine was used after its reimbursement in 2019 (Additional file 1: Fig. S4). Overall, 73 of the 320 patients with advanced PDAC were enrolled in the IITs. There were no significant differences in patient survival between those with gHRmut and those with gHRwt, regardless of whether they were enrolled in the clinical trial (P = 0.94) (Additional file 1: Fig. S5). Enrollment in the clinical trial was not prognostic significance in the multivariate Cox regression analysis (hazard ratio 1.06, 95% CI 0.72–1.58, P = 0.756) (Fig. 5).
For the sensitivity analysis, five patients were excluded due to unknown initial treatments at referral hospital in three and no treatment in two. The remaining 363 patients with advanced stage cancer were analyzed. The regimen is listed in Additional file 1: Table S5. Patients who received gemcitabine monotherapy had extremely poor outcomes, with a median OS of 5.1 months compared to those who received other regimens, whose median OS ranged from 12.0 to 15.7 months (Additional file 1: Fig. S6A). The survival benefits of 1L platinum-based chemotherapy in patients with gHRmut remained significant when all 363 stage III/IV patients were included in the analysis (Additional file 1: Fig. S6B).
Discussion
In this study, DNA from PBMC of 527 patients with PDAC were sequenced via exome sequencing, and the characteristics of 80 oncogenic candidate genes were analyzed. To our knowledge, our study is, to date, the largest PDAC cohort with comprehensive germline genetic profiling and treatment outcomes in the Asian population and may serve as a useful resource for investigating the mechanisms underlying PDAC. Overall, we identified 120 P/LP variants in 104 (19.7%) PDAC patients. Furthermore, more than half of them were high-penetrance genes. Since the prognosis of PDAC is extremely poor, and early diagnosis provides the best chance of cure nowadays, genetic counseling and cancer screening programs should be recommended to at-risk families [43].
The median age of PDAC diagnosis in our study was significantly younger in patients with a family history of BRCA-associated hereditary cancers, including breast, ovarian, prostate, and pancreatic cancer. However, there was no significant difference in the age of diagnosis between patients with or without germline mutations in the 80 oncogenic candidate genes or the 12 HR genes, suggesting that other genes, and/or environmental and lifestyle factors may also play an important role in tumorigenesis. Future studies should consider analyzing not only these 80 oncogenic candidate genes but also additional genes. However, germline variants in other genes may be rare, making it challenging to assess their impacts in a cohort of this size. A larger-scale nationwide or international collaboration program, such as the PRECEDE Consortium [44], is needed to address this issue.
In our study, 54 of 120 (45%) P/LP variants were of HR genes. The median OS was not significantly different among patients with gHRmut, other germline mutations or no mutations (P = 0.43, Fig. 4A), suggesting that gHRmut per se may not be a prognostic factor. On the other hand, patients with gHRmut who received 1L platinum-based chemotherapy had significantly better OS compared to those who received platinum-free chemotherapy. However, the median OS was not significantly different between patients with gHRwt who received 1L platinum-based versus platinum-free chemotherapy. Our study confirmed previous findings that PDAC patients with gHRmut have better responses and superior overall survival when treated with platinum-based chemotherapy [32, 45,46,47]. These findings suggest that gHRmut may serve as a predictive biomarker for platinum-based chemotherapy.
Germline BRCA1/2 mutations have been linked to inherited risks of PDAC. In total, 3.98% of patients harbored germline P/LP variants in either BRCA1 or BRCA2 in our cohort, which is similar to the results of investigations among Western (3.5%) and Japanese (3.4%) populations [13, 48]. The PARP inhibitor olaparib is approved by the U.S. Food and Drug Administration as maintenance therapy only in metastatic PDAC patients with germline BRCA1/2 mutations [5]. While germline BRCA1/2 mutation consisted of only 2–3% of PDAC patients, the broad definition of gHRmut was identified in more than 10% of PDAC patients. A previous retrospective study suggested that olaparib is also effective in patients with gHRmut other than BRCA1/2 [49]. Germline genetic profiling of all advanced PDAC patients will help to identify candidates who might benefit from PARP inhibitors. The overall incidence of gHRmut in our cohort was 9.7% (51/527), which is in line with the results of previous reports (9.7–16.5%) [32, 50,51,52]. The incidence of gHRmut in current study might be underestimated because patients with multiple primary cancers, who were more likely to harbor germline cancer susceptibility gene mutations, were excluded from initial study [53].
ATM is involved in DNA double-strand break repair and is necessary to maintain genome stability. ATM mutations cause ataxia-telangiectasia (AT) and have been associated with a risk of breast cancer [54, 55]. Yang et al. suggested that the primary types of ATM mutations are nonsense and frameshift mutations and that mutation carriers among Chinese BRCA1/2-negative breast cancer patients are more likely to have a family history of cancer [56]. In our results, ten patients (1.90%, 10/527) harbored P/LP mutations in the ATM gene, and five (50%, 5/10) carriers had a family history of cancer. ATM p.Lys468fs (40%, 4/10) was the most prevalent ATM mutation in our study, which was observed in AT patients and may cause the development of AML in children [57]. However, the effect of this frameshift mutation on the risk of pancreatic cancer remains undetermined.
Before 2019, only gemcitabine and S1 were reimbursed for advanced PDAC treatment in Taiwan. Given the limited treatment options available at that time, one of the primary objectives of our IITs was to provide relatively safe and effective treatment for them. Consequently, the inclusion criteria in the trial closely resembled those of our standard approach in daily patient care, which was reflected by the lack of a significant difference in survival between patients enrolled and not enrolled in the clinical trial. Both the GOFL and SLOG regimens were incorporated into daily practice in our hospitals prior to the reimbursement of nab-paclitaxel and FOLFORINOX [58,59,60,61,62]. Patients treated with gemcitabine monotherapy (n = 21), usually prescribed for fragile patients as recommended in the majority of treatment guidelines, were excluded in the present study to prevent selection bias when comparing survival between patients treated with platinum-based and platinum-free chemotherapy. Patients treated with other non-standard regimens (n = 20) were also excluded because only less than five patients were treated by each regimen (Additional file 1: Table S5). Although these non-standard regimens could be further divided into platinum-based or platinum-free regimens, it was unsuitable to include such a heterogeneous population in the analysis.
This study has several limitations. First, patients with a history of other cancers were excluded from this study. Germline mutations in cancer susceptibility genes not only increase the risk of PDAC but also other cancer types. Thus, possible valuable germline mutations that increase the risk of multiple cancers may be lost in our cohort. Second, we only used 80 genes associated with pancreas germline mutations based on the literature review, other oncogenic genes may also contribute to the development of pancreatic cancer. The definition of HR genes was also based on a literature review. The board definition of homologous recombination deficiency (HRD) consists of not only gene mutations but also the identification of genomic scar such as loss of heterozygosity, number of telomeric imbalances, or large-scale transitions which was not included in the current analysis [63]. As a result, some patients with HRD may have been classified as non-HRD in our cohort. While there is currently no consensus on the definition of HRD, the harmonization of an HRD definition will be a critical issue in the future. Third, the family history of patients was self-reported, which may limit the accuracy of the results. Finally, this was an observational study without treatment interventions, and genomic information was not available at the time of treatment onset. Therefore, physicians selected the first-line regimen based on the patient’s condition and preference, potentially introducing bias. This is reflected in a trend favoring the 1L platinum-based group, characterized by factors such as younger age (P = 0.091), a higher proportion in stage III than stage IV (P = 0.075), lower baseline CA19-9 levels (P = 0.072), and greater enrollment in clinical trials (P = 0.052), although these differences are not statistically significant. Furthermore, the patient number in the gHRmut cohort was very limited, comprising only 32 patients. As a result, a prospective validation clinical trial with adequate case number to determine whether gHRmut could guide the first-line treatment selection in advanced PDAC is warranted.
Conclusions
Our results identified the germline mutation profiles in Asian populations with pancreatic cancer, such mutations are important not only in the Western population but also in the Asian population. The known and/or unidentified mutations found in our study provide novel insights into the molecular pathogenesis of pancreatic cancer. Our results may help in the prevention, early diagnosis, and risk management of pancreatic cancer. Despite the limited number of cases in the current study, the observed significant survival benefits in patients with gHRmut treated with platinum-based chemotherapy are consistent with earlier reports. This highlights the importance of germline testing for all patients with advanced PDAC at the time of diagnosis.
Availability of data and materials
All data generated or analyzed during this study are available from the corresponding author on reasonable request.
Abbreviations
- ACMG:
-
American College of Medical Genetics and Genomics
- MAF:
-
Minor allele frequency
- VUS:
-
Variants of uncertain significance
- HR:
-
Homologous recombination
- IQR:
-
Interquartile range
- SLOG:
-
S1, leucovorin, oxaliplatin, and gemcitabine
- mFOLFIRINOX:
-
Folinic acid, fluorouracil, irinotecan, and oxaliplatin
- GOFL:
-
Gemcitabine, oxaliplatin, fluorouracil, and leucovorin
- Gem:
-
Gemcitabine
References
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48.
Hung YH, Chen LT, Hung WC. The trinity: interplay among cancer cells, fibroblasts, and immune cells in pancreatic cancer and implication of CD8(+) T cell-orientated therapy. Biomedicines. 2022;10(4):926.
Rahib L, Wehner MR, Matrisian LM, Nead KT. Estimated projection of US cancer incidence and death to 2040. JAMA Netw Open. 2021;4(4): e214708.
Wainberg ZA, Hochster HS, Kim EJ, George B, Kaylan A, Chiorean EG, Waterhouse DM, Guiterrez M, Parikh A, Jain R, Carrizosa DR, Soliman HH, Lila T, Reiss DJ, Pierce DW, Bhore R, Banerjee S, Lyons L, Louis CU, Ong TJ, O’Dwyer PJ. Open-label, phase I study of nivolumab combined with nab-paclitaxel plus gemcitabine in advanced pancreatic cancer. Clin Cancer Res. 2020;26(18):4814–22.
Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, Park JO, Hochhauser D, Arnold D, Oh DY, Reinacher-Schick A, Tortora G, Algul H, O’Reilly EM, McGuinness D, Cui KY, Schlienger K, Locker GY, Kindler HL. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381(4):317–27.
Quante AS, Ming C, Rottmann M, Engel J, Boeck S, Heinemann V, Westphalen CB, Strauch K. Projections of cancer incidence and cancer-related deaths in Germany by 2020 and 2030. Cancer Med. 2016;5(9):2649–56.
Fletcher O, Houlston RS. Architecture of inherited susceptibility to common cancer. Nat Rev Cancer. 2010;10(5):353–61.
Foulkes WD. Inherited susceptibility to common cancers. N Engl J Med. 2008;359(20):2143–53.
Petrucelli N, Daly MB, Pal T. BRCA1- and BRCA2-associated hereditary breast and ovarian cancer. In: Adam MP, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, Gripp KW, Amemiya A, editors. GeneReviews((R)). Seattle: University of Washington; 1993.
Klein AP, Hruban RH, Brune KA, Petersen GM, Goggins M. Familial pancreatic cancer. Cancer J. 2001;7(4):266–73.
Takai E, Yachida S, Shimizu K, Furuse J, Kubo E, Ohmoto A, Suzuki M, Hruban RH, Okusaka T, Morizane C. Germline mutations in Japanese familial pancreatic cancer patients. Oncotarget. 2016;7(45):74227.
Brand R, Borazanci E, Speare V, Dudley B, Karloski E, Peters MLB, Stobie L, Bahary N, Zeh H, Zureikat A, Hogg M, Lee K, Tsung A, Rhee J, Ohr J, Sun W, Lee J, Moser AJ, DeLeonardis K, Krejdovsky J, Dalton E, LaDuca H, Dolinsky J, Colvin A, Lim C, Black MH, Tung N. Prospective study of germline genetic testing in incident cases of pancreatic adenocarcinoma. Cancer. 2018;124(17):3520–7.
Mizukami K, Iwasaki Y, Kawakami E, Hirata M, Kamatani Y, Matsuda K, Endo M, Sugano K, Yoshida T, Murakami Y, Nakagawa H, Spurdle AB, Momozawa Y. Genetic characterization of pancreatic cancer patients and prediction of carrier status of germline pathogenic variants in cancer-predisposing genes. EBioMedicine. 2020;60: 103033.
Salo-Mullen EE, O’Reilly EM, Kelsen DP, Ashraf AM, Lowery MA, Yu KH, Reidy DL, Epstein AS, Lincoln A, Saldia A, Jacobs LM, Rau-Murthy R, Zhang L, Kurtz RC, Saltz L, Offit K, Robson ME, Stadler ZK. Identification of germline genetic mutations in patients with pancreatic cancer. Cancer. 2015;121(24):4382–8.
Gonda TA, Everett JN, Wallace M, Simeone DM, PRECEDE Consortium. Recommendations for a more organized and effective approach to the early detection of pancreatic cancer from the PRECEDE (pancreatic cancer early detection) consortium. Gastroenterology. 2021;161(6):1751–7.
Pishvaian MJ, Blais EM, Brody JR, Lyons E, DeArbeloa P, Hendifar A, Mikhail S, Chung V, Sahai V, Sohal DPS, Bellakbira S, Thach D, Rahib L, Madhavan S, Matrisian LM, Petricoin EF 3rd. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: a retrospective analysis of the know your tumor registry trial. Lancet Oncol. 2020;21(4):508–18.
Golan T, Kanji ZS, Epelbaum R, Devaud N, Dagan E, Holter S, Aderka D, Paluch-Shimon S, Kaufman B, Gershoni-Baruch R, Hedley D, Moore MJ, Friedman E, Gallinger S. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br J Cancer. 2014;111(6):1132–8.
Wattenberg MM, Asch D, Yu S, O’Dwyer PJ, Domchek SM, Nathanson KL, Rosen MA, Beatty GL, Siegelman ES, Reiss KA. Platinum response characteristics of patients with pancreatic ductal adenocarcinoma and a germline BRCA1, BRCA2 or PALB2 mutation. Br J Cancer. 2020;122(3):333–9.
Perkhofer L, Gout J, Roger E, Kude de Almeida F, Baptista Simoes C, Wiesmuller L, Seufferlein T, Kleger A. DNA damage repair as a target in pancreatic cancer: state-of-the-art and future perspectives. Gut. 2021;70(3):606–17.
Golan T, O’Kane GM, Denroche RE, Raitses-Gurevich M, Grant RC, Holter S, Wang Y, Zhang A, Jang GH, Stossel C, Atias D, Halperin S, Berger R, Glick Y, Park JP, Cuggia A, Williamson L, Wong HL, Schaeffer DF, Renouf DJ, Borgida A, Dodd A, Wilson JM, Fischer SE, Notta F, Knox JJ, Zogopoulos G, Gallinger S. Genomic features and classification of homologous recombination deficient pancreatic ductal adenocarcinoma. Gastroenterology. 2021;160(6):2119-2132.e2119.
Shan YS, Chen LT, Wu JS, Chang YF, Lee CT, Wu CH, Chiang NJ, Huang HE, Yen CJ, Chao YJ, Tsai HJ, Chen CY, Kang JW, Kuo CF, Tsai CR, Weng YL, Yang HC, Liu HC, Chang JS. Validation of genome-wide association study-identified single nucleotide polymorphisms in a case-control study of pancreatic cancer from Taiwan. J Biomed Sci. 2020;27(1):69.
Samadder NJ, Riegert-Johnson D, Boardman L, Rhodes D, Wick M, Okuno S, Kunze KL, Golafshar M, Uson PL, Mountjoy L. Comparison of universal genetic testing vs guideline-directed targeted testing for patients with hereditary cancer syndrome. JAMA Oncol. 2021;7(2):230–7.
Rio FT, Lavoie J, Hamel N, Geyer FC, Kushner YB, Novak DJ, Wark L, Capelli C, Reis-Filho JS, Mai S. Homozygous BUB1B mutation and susceptibility to gastrointestinal neoplasia. N Engl J Med. 2010;363(27):2628–37.
Kobayashi H, Ohno S, Sasaki Y, Matsuura M. Hereditary breast and ovarian cancer susceptibility genes. Oncol Rep. 2013;30(3):1019–29.
Tung N, Domchek SM, Stadler Z, Nathanson KL, Couch F, Garber JE, Offit K, Robson ME. Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat Rev Clin Oncol. 2016;13(9):581–8.
Masliah-Planchon J, Lévy D, Héron D, Giuliano F, Badens C, Fréneaux P, Galmiche L, Guinebretierre J-M, Cellier C, Waterfall JJ. Does ATRX germline variation predispose to osteosarcoma? Three additional cases of osteosarcoma in two ATR-X syndrome patients. Eur J Hum Genet. 2018;26(8):1217–21.
Casula M, Colombino M, Satta MP, Cossu A, Ascierto PA, Bianchi-Scarra G, Castiglia D, Budroni M, Rozzo C, Manca A. BRAF gene is somatically mutated but does not make a major contribution to malignant melanoma susceptibility: the Italian Melanoma Intergroup Study. J Clin Oncol. 2004;22(2):286–92.
Reznik R, Hendifar AE, Tuli R. Genetic determinants and potential therapeutic targets for pancreatic adenocarcinoma. Front Physiol. 2014;5:87.
Andersson U, Schwartzbaum J, Wiklund F, Sjöström S, Liu Y, Tsavachidis S, Ahlbom A, Auvinen A, Collatz-Laier H, Feychting M. A comprehensive study of the association between the EGFR and ERBB2 genes and glioma risk. Acta Oncol. 2010;49(6):767–75.
Rogers CD, Heijden MSVD, Brune K, Yeo CJ, Hruban RH, Kern SE, Goggins M. The genetics of FANCC and FANCG in familial pancreatic cancer. Cancer Biol Ther. 2004;3(2):167–9.
Angeli D, Salvi S, Tedaldi G. Genetic predisposition to breast and ovarian cancers: how many and which genes to test? Int J Mol Sci. 2020;21(3):1128.
Park W, Chen J, Chou JF, Varghese AM, Kenneth HY, Wong W, Capanu M, Balachandran V, McIntyre CA, El Dika I. Genomic methods identify homologous recombination deficiency in pancreas adenocarcinoma and optimize treatment selection. Clin Cancer Res. 2020;26(13):3239–47.
Park W, Chen J, Chou JF, Varghese AM, Yu KH, Wong W, Capanu M, Balachandran V, McIntyre CA, El Dika I, Khalil DN, Harding JJ, Ghalehsari N, McKinnell Z, Chalasani SB, Makarov V, Selenica P, Pei X, Lecomte N, Kelsen DP, Abou-Alfa GK, Robson ME, Zhang L, Berger MF, Schultz N, Chan TA, Powell SN, Reis-Filho JS, Iacobuzio-Donahue CA, Riaz N, O’Reilly EM. Genomic methods identify homologous recombination deficiency in pancreas adenocarcinoma and optimize treatment selection. Clin Cancer Res. 2020;26(13):3239–47.
Su YY, Chiang NJ, Chiu TJ, Huang CJ, Hsu SJ, Lin HC, Yang SH, Yang Y, Chou WC, Chen YY, Bai LY, Li CP, Chen JS. Systemic treatments in pancreatic cancer: Taiwan pancreas society recommendation. Biomed J. 2023. https://doi.org/10.1016/j.bj.2023.100696.
Ch’ang HJ, Huang CL, Wang HP, Shiah HS, Chang MC, Jan CM, Chen JS, Tien YW, Hwang TL, Lin JT, Cheng AL, Whang-Peng J, Chen LT. Phase II study of biweekly gemcitabine followed by oxaliplatin and simplified 48-h infusion of 5-fluorouracil/leucovorin (GOFL) in advanced pancreatic cancer. Cancer Chemother Pharmacol. 2009;64(6):1173–9.
Chen L-T. DL16—chemo-radiotherapy in adjuvant therapy of curatively resected pancreatic cancer: lesions from TCOG T3207 study. Ann Oncol. 2019;30:vi75.
Chiang NJ, Tsai KK, Hsiao CF, Yang SH, Hsiao HH, Shen WC, Hsu C, Lin YL, Chen JS, Shan YS, Chen LT. A multicenter, phase I/II trial of biweekly S-1, leucovorin, oxaliplatin and gemcitabine in metastatic pancreatic adenocarcinoma-TCOG T1211 study. Eur J Cancer. 2020;124:123–30.
Tsai H-J, Yang S-H, Hsiao C-F, Kao H-F, Shan Y-S, Lin Y-L, Yen C-J, Du J-S, Hsu C, Wu I-C, Chen L-T. A phase I study of biweekly abraxane in combination with oxaliplatin and oral S-1/leucovorin as first line treatment for advanced gastric, pancreatic and biliary tract cancers. Ann Oncol. 2021;32:S146–7.
Chiang N-J, Shan Y-S, Bai L-Y, Li C-P, Chen J-S, Yang S-H, Kuo Y-C, Chao Y, Hsieh Y-Y, Kao H-F, Hsiao C-F, Chen L-T. TCOG T5217 trial: a phase II randomized study of SLOG versus modified FOLFIRINOX as the first-line treatment in locally advanced or metastatic pancreatic ductal adenocarcinoma. J Clin Oncol. 2021;39(15_suppl):4143–4143.
Su YY, Chiu YF, Li CP, Yang SH, Lin J, Lin SJ, Chang PY, Chiang NJ, Shan YS, Ch’ang HJ, Chen LT. A phase II randomised trial of induction chemotherapy followed by concurrent chemoradiotherapy in locally advanced pancreatic cancer: the Taiwan Cooperative Oncology Group T2212 study. Br J Cancer. 2022;126(7):1018–26.
Bai LY, Li CP, Shan YS, Chuang SC, Chen JS, Chiang NJ, Chen YY, Tsou HH, Chuang MH, Chiu CF, Liu TW, Chen LT. A prospective phase II study of biweekly S-1, leucovorin, and gemcitabine in elderly patients with locally advanced or metastatic pancreatic adenocarcinoma—the Taiwan Cooperative Oncology Group T1217 study. Eur J Cancer. 2022;173:123–32.
Su YY, Bai LY, Shih YH, Shan YS, Chen LT. A phase II randomized study of gemcitabine and nab-paclitaxel in combination with S-1/LV (GASL) or oxaliplatin (GAP) as first-line treatment for metastatic pancreatic cancer. ClinicalTrials.gov identifier: NCT05026905. Updated Feb 08 2023. https://www.clinicaltrials.gov/ct2/show/NCT05026905. Accessed 15 Oct 2023.
Rainone M, Singh I, Salo-Mullen EE, Stadler ZK, O’Reilly EM. An emerging paradigm for germline testing in pancreatic ductal adenocarcinoma and immediate implications for clinical practice: a review. JAMA Oncol. 2020;6(5):764–71.
Katona BW, Klute K, Brand RE, Everett JN, Farrell JJ, Hawthorne K, Kaul V, Kupfer SS, Paiella S, Simeone DM, Sussman DA, Zogopoulos G, Lucas AL, Kastrinos F, PRECEDE Consortium. Racial, ethnic, and sex-based disparities among high-risk individuals undergoing pancreatic cancer surveillance. Cancer Prev Res. 2023;16(6):343–52.
Pishvaian MJ, Blais EM, Brody JR, Rahib L, Lyons E, De Arbeloa P, Hendifar A, Mikhail S, Chung V, Sohal DP. Outcomes in patients with pancreatic adenocarcinoma with genetic mutations in DNA damage response pathways: results from the know your tumor program. JCO Precis Oncol. 2019;3:1–10.
Uson PL Jr, Samadder NJ, Riegert-Johnson D, Boardman L, Borad MJ, Ahn D, Sonbol MB, Faigel DO, Fukami N, Pannala R. Clinical impact of pathogenic germline variants in pancreatic cancer: results from a multicenter, prospective, universal genetic testing study. Clin Transl Gastroenterol. 2021;12(10): e00414.
Yu S, Agarwal P, Mamtani R, Symecko H, Spielman K, O’Hara M, O’Dwyer PJ, Schneider C, Teitelbaum U, Nathanson KL. Retrospective survival analysis of patients with resected pancreatic ductal adenocarcinoma and a germline BRCA or PALB2 mutation. JCO Precis Oncol. 2019;3:1–11.
Singhi AD, George B, Greenbowe JR, Chung J, Suh J, Maitra A, Klempner SJ, Hendifar A, Milind JM, Golan T, Brand RE, Zureikat AH, Roy S, Schrock AB, Miller VA, Ross JS, Ali SM, Bahary N. Real-time targeted genome profile analysis of pancreatic ductal adenocarcinomas identifies genetic alterations that might be targeted with existing drugs or used as biomarkers. Gastroenterology. 2019;156(8):2242-2253.e2244.
Tan E, Hicks JK, Blue K, Kim RD, Carballido EM, Kim DW. Efficacy of olaparib therapy in metastatic pancreatic ductal adenocarcinoma (PDAC) with homologous recombination deficiency (HRD). J Clin Oncol. 2021;39(15_suppl): e16266.
Shui L, Li X, Peng Y, Tian J, Li S, He D, Li A, Tian B, Li M, Gao H. The germline/somatic DNA damage repair gene mutations modulate the therapeutic response in Chinese patients with advanced pancreatic ductal adenocarcinoma. J Transl Med. 2021;19(1):1–16.
Yurgelun MB, Chittenden AB, Morales-Oyarvide V, Rubinson DA, Dunne RF, Kozak MM, Qian ZR, Welch MW, Brais LK, Da Silva A. Germline cancer susceptibility gene variants, somatic second hits, and survival outcomes in patients with resected pancreatic cancer. Genet Med. 2019;21(1):213–23.
Casolino R, Paiella S, Azzolina D, Beer PA, Corbo V, Lorenzoni G, Gregori D, Golan T, Braconi C, Froeling FE. Homologous recombination deficiency in pancreatic cancer: a systematic review and prevalence meta-analysis. J Clin Oncol. 2021;39(23):2617–31.
Bychkovsky BL, Lo MT, Yussuf A, Horton C, Richardson M, LaDuca H, Garber JE, Rana HQ. Prevalence and spectrum of pathogenic variants among patients with multiple primary cancers evaluated by clinical characteristics. Cancer. 2022;128(6):1275–83.
Kurian AW, Hare EE, Mills MA, Kingham KE, McPherson L, Whittemore AS, McGuire V, Ladabaum U, Kobayashi Y, Lincoln SE. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014;32(19):2001.
Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, North B, Jayatilake H, Barfoot R, Spanova K. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38(8):873–5.
Yang Z, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, Zhang J, Xie Y. Prevalence and characterization of ATM germline mutations in Chinese BRCA1/2-negative breast cancer patients. Breast Cancer Res Treat. 2019;174(3):639–47.
Lin C-H, Lin W-C, Wang C-H, Ho Y-J, Chiang I-P, Peng C-T, Wu K-H. Child with ataxia telangiectasia developing acute myeloid leukemia. J Clin Oncol. 2010;28(14):e213-214.
Su YY, Chiang NJ, Tsai HJ, Yen CJ, Shan YS, Chen LT. The impact of liposomal irinotecan on the treatment of advanced pancreatic adenocarcinoma: real-world experience in a Taiwanese cohort. Sci Rep. 2020;10(1):7420.
Su YY, Ting YL, Wang CJ, Chao YJ, Liao TK, Su PJ, Chiang NJ, Liao IC, Yu YT, Liu YS, Tsai HM, Li YJ, Huang CJ, Liu IT, Tsai HJ, Yen CJ, Shan YS, Chen LT. Improved survival with induction chemotherapy and conversion surgery in locally advanced unresectable pancreatic cancer: a single institution experience. Am J Cancer Res. 2022;12(5):2189–202.
Su YY, Chiang NJ, Li CP, Yen CJ, Yang SH, Chou WC, Chen JS, Chiu TJ, Chen YY, Chuang SC, Bai LY, Chiu CF, Peng CM, Chan DC, Chiu SC, Yang YH, Shan YS, Chen LT. Dosing pattern and early cumulative dose of liposomal irinotecan in metastatic pancreatic cancer: a real-world multicenter study. Front Oncol. 2022;12: 800842.
Su YY, Chao YJ, Wang CJ, Liao TK, Su PJ, Huang CJ, Chiang NJ, Yu YT, Tsai HM, Chen LT, Shan YS. The experience of neoadjuvant chemotherapy versus upfront surgery in resectable pancreatic cancer: a cross sectional study. Int J Surg. 2023;109(9):2614–23.
Su YY, Chiang NJ, Yang YH, Yen CJ, Bai LY, Chiu CF, Chuang SC, Yang SH, Chou WC, Chen JS, Chiu TJ, Chen YY, Chan DC, Peng CM, Chiu SC, Li CP, Shan YS, Chen LT. Real-world data validation of NAPOLI-1 nomogram for the prediction of overall survival in metastatic pancreatic cancer. Cancers. 2023;15(4):1008.
Stewart MD, Merino Vega D, Arend RC, Baden JF, Barbash O, Beaubier N, Collins G, French T, Ghahramani N, Hinson P, Jelinic P, Marton MJ, McGregor K, Parsons J, Ramamurthy L, Sausen M, Sokol ES, Stenzinger A, Stires H, Timms KM, Turco D, Wang I, Williams JA, Wong-Ho E, Allen J. Homologous recombination deficiency: concepts, definitions, and assays. Oncologist. 2022;27(3):167–74.
Acknowledgements
Not applicable.
Funding
This work was supported by the Ministry of Science and Technology, Taiwan [MOST 110-2321-B-037-002 To D.Y.H] [MOST 111-2314-B-400-027 to L.T.C], the Kaohsiung Medical University, Taiwan [KMU-KI110001] and National Health Research Institutes, Taiwan [CA-112-PP-07].
Author information
Authors and Affiliations
Contributions
YSS, DYH, LTC designed and supervised the study. SMC, CYC and YJL performed the experiments and collected the data. NJC, YJC, CJY, CJW, CJH, HJT, SHC and SCC collected clinical information, samples and treated patients. CRT and SMC designed and collected the questionnaire. SMC and YYS performed data analysis. SMC and YYS interpreted the data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The research protocol was approved by the Ethics Committee of the National Health Research Institute (approval number EC-1030109-E), National Cheng Kung University Hospital (approval number B-BR-102-070) and Kaohsiung Medical University Hospital (approval number KMUHIRB-F(I)-20160012). Written informed consent was obtained from all patients, and all clinical investigations were conducted according to the principles of the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Median overall survival of 527 PDAC patients with or without germline gene mutations in different stage. Figure S2. Median overall survival of 107 stage III PDAC patients with or without conversion surgery. Figure S3. Timeline comparison between the investigator-initiated trials for pancreatic cancer in Taiwan and the corresponding global phase III trials. Figure S4. The distribution of the most commonly used regimen by year. An arrow indicates the time of nab-paclitaxel reimbursement in Taiwan. Figure S5. Median overall survival of 320 PDAC patients with or without enrollment in clinical trials. Figure S6. (A) Median overall survival of 363 stage III/IV PDAC patients treated by different regimens. (B) Median overall survival of 363 stage III/IV PDAC patients with or without HR gene mutations treated with 1L platinum-based or non-platinum-based chemotherapy. Table S1. Categories of the 80 candidate cancer-associated genes analyzed for germline mutations in PDAC patients. Genes that have overlapping properties are listed only once and were classified as low-, moderate-, high- and recessive penetrance. Table S2. Pathogenic and likely pathogenic germline variants in susceptibility genes. Table S3. Family history of cancer in 527 PDAC patients. Table S4. Demographics of 32 patients with germline mutation in 12 homologous recombination genes treated with first-line platinum chemotherapy or non-platinum chemotherapy. Table S5. List of all regimen used in 368 stage III/IV PDAC patients.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Cheng, S.M., Su, YY., Chiang, NJ. et al. Germline mutations of homologous recombination genes and clinical outcomes in pancreatic cancer: a multicenter study in Taiwan. J Biomed Sci 31, 21 (2024). https://doi.org/10.1186/s12929-024-01008-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12929-024-01008-7