Abstract
Background
Aerobically-grown bacteria can be challenged by hydrogen peroxide stress from endogenous aerobic metabolism and exogenously generated reactive oxygen species. Catalase (Kat), alkyl hydroperoxidase (Ahp), and glutathione peroxidase (Gpx) systems are major adaptive responses to H2O2 stress in bacteria. Stenotrophomonas maltophilia is a ubiquitous Gram-negative bacterium equipped with four Kats (KatA1, KatA2, KatMn, and KatE), one Ahp (AhpCF), and three Gpxs (Gpx1, Gpx2, and Gpx3). Here, we systematically investigated how the eight H2O2 scavenging genes differentially contribute to the low-micromolar levels of H2O2 generated from aerobic metabolism and high-millimolar levels of H2O2 from exogenous sources.
Methods
Gene expression was assessed and quantified by reverse transcription-PCR (RT-PCR) and real time quantitative PCR (qRT-PCR), respectively. The contribution of these enzymes to H2O2 stress was assessed using mutant construction and functional investigation.
Results
Of the eight genes, katA2, ahpCF, and gpx3 were intrinsically expressed in response to low-micromolar levels of H2O2 from aerobic metabolism, and the expression of katA2 and ahpCF was regulated by OxyR. AhpCF and KatA2 were responsible for alleviating aerobic growth-mediated low concentration H2O2 stress and AhpCF played a critical role for stationary-phase cells. KatA2 was upregulated to compensate for AhpCF in the case of ahpCF inactivation. After exposure to millimolar levels of H2O2, katA2 and ahpCF were upregulated in an OxyR-dependent manner. KatA2 was the critical enzyme for dealing with high concentration H2O2. Loss-of-function of KatA2 increased bacterial susceptibility to high concentration H2O2.
Conclusions
AhpCF and KatA2 are key enzymes protecting S. maltophilia from hydrogen peroxide stress.
Similar content being viewed by others
Background
In aerobic bacteria, hydrogen peroxide (H2O2) stress is endogenously generated by aerobic metabolism. Exogenous H2O2 stress can be generated by chemical processes, competing organisms, and host cells in the environment. Superoxide, H2O2, and hydroxyl radicals are three main reactive oxygen species (ROS) in aerobic bacteria. Unlike superoxide and hydroxyl radicals, H2O2 is not a free radical and is less toxic to bacteria. However, distinct from superoxide and hydroxyl radicals, H2O2 can easily diffuse across cell membranes. Furthermore, hydroxyl radical is the most reactive ROS species and it can be readily generated from H2O2 in the presence of Fe2+ via the Fenton reaction, causing irreversible damage to bacteria [1]. Therefore, effective removal of H2O2 is critical for bacterial survival.
To prevent H2O2-mediated damage, aerobic bacterial pathogens must quickly convert H2O2 into other, less dangerous substances. The most common and efficient systems for bacteria to alleviate H2O2 stresses are an array of scavenging enzymes [2], including catalase (Kat), glutathione peroxidase (Gpx), and alkyl hydroperoxidase/alkyl hydroperoxide reductase (Ahp) [3]. Catalase directly catalyzes the decomposition of hydrogen peroxide without oxidizing the enzyme itself. Peroxidases detoxify H2O2 by oxidizing itself and rely on cellular reductants to revive them from the oxidized state. A bacterium can harbor an array of H2O2 scavenging enzymes, like KatG, KatE, AhpCF, and BtuE in E. coli [4], and KatA, KatB, KatC, AhpA, AhpB, AhpCF, and BtuE in P. aeruginosa [5]. The H2O2 scavenging enzymes may differentially function in response to different oxidative stress sources.
OxyR, a LysR family transcription factor, is a well-characterized regulator of the H2O2 response in Gram-negative bacteria [6]. OxyR contains a regulatory domain and a DNA binding domain. After sensing a H2O2 threat, OxyR undergoes secondary structure rearrangement by forming a disulfide bond between the two conserved cysteine residues in the regulatory domain, resulting in oxidized OxyR. The oxidized OxyR binds to the promoter region of the target gene via the DNA binding domain, modulating target gene expression as a transcriptional activator or repressor.
Stenotrophomonas maltophilia is an aerobic, Gram-negative, γ-proteobacterium that is widely distributed in the soil, water, plant rhizosphere, and hospital equipment [7]. It is also a pathogen that infects cystic fibrosis and immunocompromised patients [8]. Because of its diverse habitats, S. maltophilia is expected to be equipped with more effective H2O2 alleviation systems to adapt to different environmental niches. Analysis of the S. maltophilia genome sequence indicates the presence of many H2O2 scavenging enzymes, including four distinct Kats, three Gpxs, and one alkyl hydroperoxidase/alkyl hydroperoxide reductase system (AhpCF) [9]. Given that three systems contribute to neutralize H2O2 stresses, a defect in a single system can be compensated by the others. Therefore, a global investigation of the three systems, instead of focusing on one system, is likely to contribute more to our understanding of H2O2 detoxification in bacteria. To our knowledge, no previous studies have comprehensively investigated the function and interplay among the three antioxidant systems in S. maltophilia. This study aimed to provide this information and elucidate the role of these antioxidant enzymes in protecting bacteria against H2O2 stress from aerobic metabolism or exogenous sources.
Methods
Bacterial strains, plasmid, and growth condition
Table S1 lists the bacterial strains, plasmids, and PCR primers used in this study. All primers used in this study were designed based on the genome of S. maltophilia K279a.
Construction of in-frame deletion mutants
The strategy of two-step double cross-over homologous recombination was used for the construction of mutants used in this study. Two PCR amplicons, corresponding to upstream and downstream of the gene intended to delete, were amplified using the paired primer sets and subsequently cloned into pEX18Tc to yield the recombinant plasmids for mutants construction. The primer sets used are KatA1N-F/KatA1N-R and KatA1C-F/KatA1C-R for plasmid pΔKatA1, KatA2N-F/KatA2N-R and KatA2C-F/KatA2C-R for plasmid pΔKatA2, KatMnN-F/KatMnN-R and KatMnC-F/KatMnC-R for plasmid pΔKatMn, KatEN-F/KatEN-R and KatEC-F/KatEC-R for plasmid pΔKatE, AhpCN-F/AhpCN-R and AhpFC-F/AhpFC-R for plasmid pΔAhpCF, Gpx1N-F/Gpx1N-R and Gpx1C-F/Gpx1C-R for plasmid pΔGpx1, Gpx2N-F/Gpx2N-R and Gpx2C-F/Gpx2C-R for plasmid pΔGpx2, and Gpx3N-F/Gpx3N-R and Gpx3C-F/Gpx3C-R for plasmid pΔGpx3 (Table S1). These pEX18Tc-derived plasmids were mobilized into KJ cells by conjugation and the transconjugants selection were performed as descried previously [10]. PCR and DNA sequencing were performed to confirm the correctness of mutants. Double, quadruple, and hepta mutants were constructed from single mutants by the same procedure.
Construction of complementation plasmids pAhpCF and pKatA2
The 2551-bp PCR amplicon containing intact ahpCF genes was obtained by PCR using the primer sets AhpCF-F and AhpCF-R and cloned into pRK415, yielding pAhpCF. An approximate 2.1-kb DNA fragment containing intact katA2 gene was obtained by PCR using primer sets KatA2N-F and KatA2C-R and cloned into pRK415, generating plasmid pKatA2.
Dihydrochodamine 123 (DHR123) assay
Overnight cultures were subcultured to fresh LB medium containing 0.9 μg/ml DHR123 with an initial OD450 of 0.15. After a 5-h and 24-h incubations, fluorescence was detected using 500 nm as the excitation wavelength and 550 nm as the emission wavelength.
Reverse transcription-PCR (RT-PCR)
The DNA-free RNA of logarithmic-phase S. maltophilia cells was extracted using Total RNA Extraction Kit Mini (ARROWTEC) and reverse transcribed to cDNA by High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The cDNA of 100 ng was used as template for PCR with the primers indicated. The primer sets used were KatA1Q-F/R for katA1, KatA2Q-F/R for katA2, KatMnQ-F/R for katMn, KatEQ-F/R for katE, AhpCQ-F/R for ahpC, Gpx1Q-F/R for gpx1, Gpx2Q-F/R for gpx2, and Gpx3Q-F/R for gpx3 (Table S1). PCR amplicons were visualized by agarose gel electrophoresis. To check the specificity of primer pairs, control PCRs were performed using the chromosome DNA as the template. Since smeX in S. maltophilia KJ is intrinsically quiescent [11], it was used as the negative control to assure RNA purity.
Real time quantitative PCR (qRT-PCR)
The cDNA prepared for the aforementioned RT-PCR assay was used as template for qRT-PCR. qRT-PCR was carried out by the ABI Prism 7000 Sequence Detection System (Applied Biosystems) according to the manufacturer’s protocols. The 16 s rRNA gene was used as an internal control and the transcripts of genes assayed were normalized with the internal control using ΔΔCT method [12]. Primers used for qRT-PCR were the same as those used for RT-PCR (Table S1). All experiments were performed in triplicate.
Construction of promoter-xylE transcriptional fusion reporter plasmids
Three DNA segments upstream and including the start codons of gpx3, katA2, and ahpC were amplified by PCR using the primer sets of Gpx3N-F/Gpx3N-R, KatA2N-F/KatA2N-R, and AhpCN-F/AhpCN-R, respectively (Table S1). These PCR products were inserted into pRKxylE to place the amplicons upstream of xylE, which encodes an enzyme with C23O activity. These plasmids were referred to as pGpx3xylE, pKatA2xylE, and pAhpCxylE, respectively.
Determination of C23O activity
Catechol 2, 3-dioxygenase (C23O), encoded by a xylE gene, catalyzes the hydrolysis of catechol into the yellow 2-hydroxymuconate semialdehyde, which can be quantitatively determined by spectrophotometric analysis at a wavelength of 375 nm. C23O activity were determined spectrophotometrically at 375 nm as described previously [11]. The rate of hydrolysis was calculated by using 44,000 M− 1 cm− 1 as the extinction coefficient. One unit of enzyme activity (U) was defined as the amount of C23O that converts 1 nmole catechol per min. The C23O specific activity was expressed as U/OD450nm.
Growth kinetic assay
Overnight-cultured strain tested was inoculated into fresh LB medium at the initial OD450nm of 0.15. The OD450nm readings were taken at interval of 3 h for a total time of 24 h.
H2O2 susceptibility test (disk diffusion assay)
The strain tested was cultured to mid-log phase and adjusted to a concentration of 107 cells/ml. A 100 μl aliquot was spread evenly over the surface of a LB agar plates. A 10 μl of 20% H2O2 was spotted onto a sterile paper disk (6 mm in diameter) and the disk was placed on the center of plate. The diameter of growth inhibition zone around disk was measured after a 24-h incubation at 37 °C.
Results
Analysis of Kat, AhpC, and Gpx systems in S. maltophilia genome
The catalase (Kat), alkyl hydroperoxidase/alkyl hydroperoxide reductase (AhpCF), and glutathione peroxidase (Gpx) systems are three major and extensively reported enzymatic H2O2 elimination systems in several bacteria. Genome sequence analysis showed that four kat, one ahpCF, and three gpx genes existed in the genome of S. maltophilia K279a [9]: Smlt0372 (katA1), Smlt1385 (katA2), Smlt2537 (katMn), Smlt3583 (katE), Smlt0841–0840 (ahpCF), Smlt3183 (gpx1), Smlt3228 (gpx2), and Smlt4676 (gpx3). In this study, we aimed to assess the roles of the eight enzymes in alleviating hydrogen peroxide stress generated by endogenous aerobic metabolism or by exogenous sources.
AhpCF and KatA2 contribute to scavenge micromolar H2O2, and AhpCF play a critical role for stationary-phase cells
The intrinsic expression of the H2O2 scavenging enzyme genes was tested using reverse transcription-PCR (RT-PCR). Of the eight genes tested, gpx3, katA2, and ahpC transcripts were detected (Fig. 1a), suggesting that Gpx3, KatA2, and AhpCF may participate in the alleviation of endogenous H2O2 stress arising from bacterial aerobic metabolism. The expressions of gpx3, katA2, and ahpC genes in the logarithmic and stationary phases were further assessed by qRT-PCR. The ahpC expression was abundant compared to katA2 and gpx3 in logarithmic phase. The expression level of ahpC was further increased in the stationary phase (Fig. 1b). These observations suggested a critical role for ahpC in endogenous H2O2 stress alleviation.
Roles of four catalases (KatA1, KatA2, KatE, and KatMn), one alkyl hydroperoxidase (AhpC), and three glutathione peroxidases (Gpx1, Gpx2, and Gpx3) in the alleviation of endogenous hydrogen peroxide stress. Bars represent the average values from three independent experiments. Error bars represent the standard error of the mean. *, P < 0.001, significance calculated by Student’s t test. (a) Agarose gel electrophoresis of reverse transcription PCR (RT-PCR). Overnight-cultured S. maltophilia KJ was inoculated into fresh LB with an initial OD450nm of 0.15 and grown for 5 h. The cDNAs were obtained using reverse transcription with random primers and PCR was performed using primer pairs (Table S1) targeting candidate genes. The smeX gene, which is not expressed in strain KJ, is used as a control for DNA contamination during cDNA preparation. (b) The expression of gpx3, katA2, and ahpC genes in logarithmic- and stationary-phase wild-type KJ cells. Overnight culture of KJ cells was inoculated into fresh LB with an initial OD450nm of 0.15. Cells were grown aerobically for 5 h or 15 h before measuring gpx3, katA2, and ahpC transcripts using qRT-PCR. All values were normalized to gpx3 transcript of logarithmic-phase KJ cells. (c) Regulatory role of OxyR in the intrinsic expression levels of gpx3, katA2, and ahpC genes. Overnight cultures of KJ and KJΔOxyR cells were inoculated into fresh LB with an initial OD450nm of 0.15. Cells were grown aerobically for 5 h or 15 h before measuring gpx3, katA2, and ahpC transcripts using qRT-PCR. All values were normalized to the transcript of logarithmic-phase KJ cells. (d) Regulatory role of OxyR in the intrinsic expression levels of gpx3, katA2, and ahpC genes. Overnight cultures of bacteria cells (KJ (Gpx3xylE), KJ (pKatA2xylE), KJ (pAhpCxylE), KJΔOxyR (Gpx3xylE), KJΔOxyR (pKatA2xylE), and KJΔOxyR (pAhpCxylE)) were inoculated into fresh LB with an initial OD450nm of 0.15. Cells were grown aerobically for 5 h or 15 h before measuring the C23O activity. All values were normalized to the activity in KJ cells. (e) Functions of OxyR, Gpx, Kat, and AhpCF systems in response to endogenously aerobic metabolism-derived H2O2 stress. The growth curves of KJ and its derived isogenic mutants were measured by reading OD450 at the time points as indicated. *, the growth difference of KJΔAhpCF and KJΔAhpCF (pAhpCF) at the 24-h time point was significant. (f) DHR 123 assay of wild-type KJ and mutants KJΔGpx3, KJΔKatA2, and KJΔAhpCF. The bacterial cells tested were cultured in LB medium containing DHR 123 for 5 h and 24 h, respectively, and the fluorescence at 550 nm was determined. The relative fluorescence is normalized to the fluorescence of wild-type KJ. (g) The expression levels of gpxs, kats, and ahpCF of KJΔGpx3, KJΔKatA2, and KJΔAhpCF in response to endogenously aerobic metabolism-derived H2O2 stress. Bacteria cultured overnight (KJ, KJΔGpx3, KJΔKatA2, and KJΔAhpC) were inoculated into fresh LB with an initial OD450nm of 0.15 and grown for 5 h. The katA1, katA2, katMn, katE, ahpC, gpx1, gpx2, and gpx3 transcripts were measured using qRT-PCR. The relative transcription level for each gene was expressed as the ratio of mutant to wild-type
OxyR is a well-known regulator response to H2O2 stress in several bacteria [13]. The regulatory role of OxyR in the intrinsic expression of gpx3, katA2 and ahpC was assessed by qRT-PCR. The expression of gpx3 was little affected by OxyR. The katA2 transcript was obviously decreased in the oxyR null mutant, indicating that OxyR is a positive regulator for the intrinsic expression of katA2. Nevertheless, OxyR acted as a repressor for the expression of ahpC in aerobically-grown cells (Fig. 1c). This observation is peculiar since OxyR is a positive regulator of antioxidant system widely reported in several bacteria [13, 14]; thus we used promoter-xylE transcriptional fusion construct to recheck the role of OxyR in the expression of gpx3, katA2, and ahpC. The same conclusion was obtained from the results of promoter-xylE transcriptional fusion assay (Fig. 1d). To investigate the roles of gpx3, katA2, and ahpCF in the alleviation of endogenously aerobic metabolism-derived H2O2 stress, we investigated the aerobic growth of different single mutants (KJΔGpx3, KJΔKatA2, and KJΔAhpCF) and different combinations of double mutants (KJΔGpx3ΔKatA2 and KJΔGpx3ΔAhpCF). After several tries, we could not successfully obtain the double mutant of katA2 and aphCF genes. In addition, KJΔOxyR was also included. None of the tested mutants showed any observable growth restrictions in the logarithmic phase. However, ahpCF-associated mutants (KJΔAhpCF and KJΔGpx3ΔAhpCF) exhibited gradual reduction of cell density in the stationary phase, and this compromise was not observed when ahpCF genes were complemented (Fig. 1e).
To assess the relatedness of deletion mutant phenotypes to the intracellular H2O2 concentrations, the intracellular H2O2 concentrations of wild-type KJ and mutants KJΔGpx3, KJΔKatA2, and KJΔAhpCF in the logarithmic (5 h) and the stationary phases (24 h) were determined by dihydrochodamine 123 (DHR123) assay. DHR123 is used for the detection of intracellular ROS and can detect H2O2 in the presence of endogenous peroxidases. The presence of ROS oxidizes DHR123 to the fluorescent derivative rhodamine 123. Thus, the intracellular H2O2 concentration is proportional to the fluorescence intensity. The fluorescences detected from the logarithmic-phase KJΔGpx3, KJΔKatA2, and KJΔAhpCF, and from the stationary-phase KJΔGpx3 and KJΔKatA2 were comparable to that from wild-type KJ (Fig. 1f). Nevertheless, stationary-phase KJΔAhpCF cells had higher fluorescence relative to stationary-phase KJ cells (Fig. 1f), correlated well with stationary-phase growth compromise of ahpCF-associated mutants (Fig. 1e).
Given functional redundancy in these H2O2-alleviating enzymes, we considered the possibility that some of these enzymes may be induced to compensate for the absence of one. To test this hypothesis, the transcription levels of the eight genes were measured using qRT-PCR in the deletion mutants KJΔGpx3, KJΔKatA2, and KJΔAhpCF. Inactivation of gpx3 or katA2 alone did not significantly affect the expression of the other seven genes. However, the expression of katA2 in KJΔAhpCF cells increased by 19 ± 2-fold compared to parental KJ cells (Fig. 1g).
KatA2 and AhpCF, mainly KatA2, contribute to scavenge millimolar H2O2
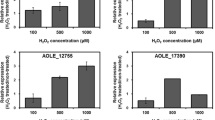
The impact of exogenous H2O2 stress on the expression of H2O2 scavenging enzymes was investigated by qRT-PCR. Of the eight genes assessed, katA2 and ahpC were upregulated after a 2 mM H2O2 challenge (Fig. 2a).
Roles of four catalases (KatA1, KatA2, KatE, and KatMn), one alkyl hydroperoxidase (AhpC), and three glutathione peroxidases (Gpx1, Gpx2, and Gpx3) in the alleviation of exogenous hydrogen peroxide stress. Bars represent the average values from three independent experiments. Error bars represent the standard error of the mean. *, P < 0.001, significance calculated by Student’s t test. (a) Expression of H2O2-hydrolyzing enzyme genes in the strains KJ, KJΔKatA2, and KJΔAhpC after hydrogen peroxide challenge. The bacteria tested were treated with 2 mM H2O2 for 10 min before measuring katA1, katA2, katMn, katE, ahpC, gpx1, gpx2, and gpx3 transcription using qRT-PCR. All values were normalized to individual transcripts obtained from untreated KJ cells. (b) Regulatory role of OxyR in katA2 expression in response to exogenous H2O2 stress. The KJ and KJΔOxyR cells were untreated or treated with different H2O2 concentration as indicated for 10 min before measuring katA2 transcript using qRT-PCR. All values were normalized to katA2 transcript obtained from untreated KJ cells. (c) Regulatory role of OxyR in ahpC expression in response to exogenous H2O2 stress. The KJ and KJΔOxyR cells were untreated or treated with different H2O2 concentration as indicated for 10 min before measuring ahpC transcript using qRT-PCR. All values were normalized to ahpC transcript obtained from untreated KJ cells. (d) H2O2 susceptibility test of KJ and its derived isogenic mutants. The bacterial cell suspension tested was uniformly spread onto MH agar, and a sterile filter paper with 10 μl of 20% H2O2 was placed on the agar. After a 24-h incubation at 37 °C, the growth inhibition zone was measured
We also assessed the possibility of compensatory expression in KJΔKatA2 and KJΔAhpCF in the presence of exogenous H2O2 stress. In either KJΔKatA2 or KJΔAhpCF, the expression levels of the remaining seven H2O2 scavenging enzymes in response to H2O2 challenge were hardly affected compared to that in wild-type KJ (Fig. 2a).
We investigated the regulatory role of OxyR in exogenous H2O2-mediated katA2 and ahpC upregulation with the H2O2 concentrations ranged from 0 to 2 mM. When the exogenous H2O2 concentration was as low as 1 μM, there was no impact on the amounts of katA2 and ahpC transcripts. In response to 5 μM or 100 μM H2O2 challenge, katA2 transcript had a mild (approximately 2–3 fold), but not significant increment; however, ahpC transcript was upregulated (Fig. 2b & c). When the challenged H2O2 concentration was higher than 250 μM, the katA2 and ahpC transcripts were significantly increased (Fig. 2b & c). In addition, katA2 expression was positively regulated by OxyR without or with the treatment of H2O2 (Fig. 2b). However, OxyR regulatory role in ahpC expression was H2O2 concentration dependent, as a repressor when H2O2 concentration was less than 5 μM and as an activator when H2O2 concentration was higher than 100 μM (Fig. 2c).
To investigate the role of the eight enzymes in exogenous H2O2 detoxification, we performed an H2O2 susceptibility test of KJ-derived mutants containing single deletions of the katA1, katA2, katMn, katE, ahpCF, gpx1, gpx2, and gpx3 genes. In addition, we assessed the H2O2 susceptibility of KJΔOxyR. Except for KJΔKatA2 and KJΔOxyR, the remaining seven mutants displayed H2O2 susceptibility that was similar to wild-type KJ (Fig. 2d). KJΔKatA2 was more sensitive to H2O2 than wild-type KJ (Fig. 2d), and complementation of the mutant with pKatA2, a plasmid containing an intact katA2 gene, restored H2O2 resistance (Fig. 2d). KJΔOxyR was also more sensitive to H2O2 than wild-type KJ, but not as severe as KJΔKatA2 (Fig. 2d). Next, we assessed whether additional mutations in KJΔKatA2 could enhance H2O2 sensitivity by constructing several combinations of multiple genes deletion mutants using KJΔKatA2 as a parental strain and performing H2O2 sensitivity assays in all mutants. H2O2 sensitivity was hardly augmented compared to KJΔKatA2 in all mutants tested, although 4 catalase genes and three gpx genes were simultaneously inactivated (KJΔ4KatΔ3Gpx) (Fig. 2d).
It has been reported that OxyR of E. coli binds to the 5′ promoter-operator regions of target genes at a conserved motif comprised of four ATAG elements spaced at 10-bp intervals [15, 16]. Since OxyR is involved in the H2O2-induced upregulation of katA2 and ahpCF, we surveyed the upstream region of the ahpCF and katA2 genes. We discovered ATAG-N14-ATAG and ATAG-N19-ATAG elements near the ahpCF and katA2 promoters (Fig. S1).
Discussion
H2O2 stress is an inevitable challenge for aerobic bacteria. Respiratory bursts account for up to 87% of the total H2O2 production in aerobically-grown Escherichia coli, and intracellular H2O2 from aerobic metabolism normally remains at low-micromolar ranges (< 4 μM) [17]. In the course of infection, H2O2 levels can reach up to millimolar concentrations because of the oxidative burst generated by host immune cells [2]. To avoid H2O2 toxicity, bacteria have equipped themselves with several scavenging enzymes to maintain intracellular H2O2 at nanomolar concentrations [4, 17]. AhpCF and catalase systems are scavenging enzymes that are extensively conserved in several bacterial lineages [2]. AhpCF is more kinetically efficient than catalases at scavenging H2O2, but its activity is more easily saturated than that of catalases [4]. Therefore, AhpCF is the primary scavenger when H2O2 is in the low-micromolar range, and catalase activity predominates when the cell reaches millimolar levels of H2O2 [4]. This paradigm has been observed in a variety of organisms [4], and we highlight our findings in this study to add new evidence to this paradigm.
AhpCF of S. maltophilia was expressed in the logarithmic phase and further upregulated in the stationary phase (Fig. 1b), implying that higher AhpCF activity is required for S. maltophilia to deal with H2O2 stress in the stationary phase. This inference is supported by the observation in Fig. 1e and Fig. 1f, since ahpCF-associated mutants (KJΔAhpCF and KJΔGpx3ΔAhpCF) exhibited compromised stationary-phase growth (Fig. 1e) and the higher H2O2 concentration was observed in the stationary-phase KJΔAhpCF cells (Fig. 1f). Inactivation of katA2 did not affect the expression of other H2O2 scavenging enzymes (Fig. 1g) and did not compromise bacterial aerobic growth (Fig. 1e), indicating that AhpCF alone is potent enough to deal with the low-micromolar H2O2 stress. In contrast, upregulation of KatA2 is needed to attain normal logarithmic growth in the case of ahpCF inactivation (KJΔAhpCF) (Fig. 1e and g). Collectively, for an aerobically-grown S. maltophilia, AhpCF and KatA2 are key enzymes responsible for the alleviation of logarithmic-phase H2O2 stress and AhpCF system plays a critical role in dealing with the stationary-phase H2O2 stress.
When bacteria encounter exogenous H2O2 stress up to the high-micromolar, even millimolar level, ahpCF and katA2 are upregulated (Fig. 2a), linking the contribution of AhpCF and KatA2 to alleviate high concentration H2O2. However, neither KJΔKatA2 nor KJΔAhpCF exhibited compensatory expression of other enzymes tested in response to 2 mM H2O2 challenge (Fig. 2a), suggesting that there should be other non-enzymatic systems contributing to deal with millimolar H2O2 stress in addition to KatA2 and AhpCF. However, we also observed that the katA2-associated mutants, but not the other mutants, had a compromised H2O2 tolerance (Fig. 2d), indicating that among the enzymes tested in this study, KatA2 is the dominant enzyme for the alleviation of high concentration H2O2 stress.
Vattanaviboon’s group has investigated the role of AhpCF of S. maltophilia in response to high level of H2O2 stress recently [18], and their conclusions are not totally consistent with our findings. They demonstrated that inactivation of ahpC rendered S. maltophilia more resistant to 300–900 mM H2O2 than parental strain, which was attributed to the enhanced KatA2 expression and activity [18]. However, our results showed that the expression of katA2 in the 2 mM H2O2-treated ahpCF mutant (KJΔAhpCF) was comparable to that of parental strain (KJ) (Fig. 2a). The discrepancy may be attributed to different stress intensities (the treated H2O2 concentration and time intervals), different experiment designs for H2O2 tolerance evaluation, and strain variation. If the ahpC mutant indeed gains a survival superiority against H2O2 at concentrations commonly used in a hospital, the prevalence of ahpC mutant in the clinical S. maltophilia isolates should be an interesting issue to study.
The OxyR regulatory role is another interesting finding in this study. OxyR is an H2O2-sensing transcriptional regulator that is generally conserved in Gram-negative bacteria [13, 14]. In this study, H2O2 dose-dependent regulation was observed in S. maltophilia OxyR. OxyR functioned as a positive regulator for the expression of katA2 either at micromolar or at millimolar H2O2 concentrations (Fig. 1c, d, & b). However, OxyR played a double-edged role in the regulation of ahpCF expression. OxyR repressed ahpCF expression at low-micromolar H2O2 concentrations (H2O2 concentration < 5 μM) (Fig. 1c, d & c), but activated ahpCF expression when H2O2 H2O2 concentration higher than 100 μM (Fig. 2c). This is uncommon because OxyR generally promotes ahpCF expression in other bacteria [19]. Herein, we proposed two possibilities to explain this observation. (i) Two different OxyR activated forms may form dependent on the H2O2 concentrations (different symbols for active OxyR in Fig. 3a and b), which may have different impacts on ahpCF expression (Fig. 3). (ii) Members of OxyR regulon triggered by low H2O2 concentrations are not totally the same as those triggered by high H2O2 concentrations, and different OxyR regulon member(s) regulate(s) the ahpCF expression in micromolar and millimolar H2O2 concentrations, respectively (Fig. 3a and b). The negative regulatory role of OxyR in ahpCF expression (Fig. 1c, d) may help S. maltophilia to cope with the endogenous H2O2 stress in the case of the loss of OxyR function. When oxyR is inactivated, the shortage of KatA2 activity can be compensated by upregulated AhpCF, which can maintain normal H2O2 detoxification. This may be the reason why KJΔOxyR displayed comparable growth with wild-type KJ, but KJΔAhpCF had a growth compromise in the stationary phase (Fig. 1e).
A model for H2O2-dependent and OxyR-mediated transcription regulation of ahpCF and katA2 genes in response to different concentrations of H2O2 stress in S. maltophilia. (a) Low-micromolar H2O2 is generated by bacterial aerobic metabolism and OxyR is oxidized at a specific “sensing” cysteine residue by H2O2. The activated OxyR represses the expression of ahpCF operon and increases the expression of katA2 gene, either directly or indirectly. (b) When bacteria encounter exogenous H2O2 stress and the intracellular H2O2 concentration increases to millimolar levels, activated OxyR activates the expression of ahpCF operon and katA2 gene, either directly or indirectly
Conclusion
AhpCF and KatA2 are two main enzymes to differentially protect S. maltophilia from the hydrogen peroxide stress. AhpCF and KatA2 participate the alleviation of low-micromolar level H2O2, and AhpCF has a crucial role for stationary-phase cells; in contrast, KatA2 is the major contributor for dealing with the millimolar level H2O2. OxyR acts as a positive regulator for the expression of katA2. However, the regulatory role of OxyR in the ahpCF expression depends on the H2O2 concentration, as a repressor in H2O2 of low-micromolar level and as an activator in H2O2 of millimolar level.
Availability of data and materials
Data and materials related to this study are available upon request.
Abbreviations
- Ahp:
-
Alkyl hydroperoxidase
- DHR123:
-
Dihydrochodamine 123
- Gpx:
-
Glutathione peroxidase
- Kat:
-
Catalase
- QRT-PCR:
-
Real time quantitative PCR
- ROS:
-
Reactive oxygen species
- RT-PCR:
-
Reverse transcription-PCR
References
Cabiscol E, Tamarit J, Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol. 2000;3:2–8.
Mishra S, Imlay J. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem Biophys. 2012;525:145–60.
Poole LB. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys. 2005;433:240–54.
Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001;183:7173–81.
Ochsner UA, Vasil ML, Alsabbagh E, Parvatiyar K, Hassett DJ. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J Bacteriol. 2000;182:4533–44.
Demple B. Regulation of bacterial oxidative stress genes. Annu Rev Genet. 1991;25:315–37.
Alavi P, Starcher MR, Thallinger GG, Zachow C, Muller H, Berg G. Stenotrophomonas comparative genomics reveals genes and functions that differentiate beneficial and pathogenic bacteria. BMC Genomics. 2014;15:482.
Parkins MD, Floto RA. Emerging bacterial pathogens and changing concepts of bacterial pathogenesis in cystic fibrosis. J Cyst Fibros. 2015;14:293–304.
Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, et al. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Gen Biol. 2008;9:R74.
Yang TC, Huang YW, Hu RM, Huang SC, Lin YT. AmpDI is involved in expression of the chromosomal L1 and L2 β-lactamases of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2009;53:2902–7.
Chen CH, Huang CC, Chung TC, Hu RM, Huang YW, Yang TC. Contribution of resistance-nodulation-division efflux pump operon smeU1-V-W-U2-X to multidrug resistance of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2011;55:5826–33.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(Delta Delta C(T)) method. Methods. 2001;25:402–8.
Dubbs JM, Mongkolsuk S. Peroxide-sensing transcriptional regulators in bacteria. J Bacteriol. 2012;194:5495–503.
Chen H, Xu G, Zhao Y, Tian B, Lu H, Yu X, Xu Z, Ying N, Hu S, Hua Y. A novel OxyR sensor and regulator of hydrogen peroxide stress with one cysteine residue in Deinococcus radiodurans. PLoS One. 2008;3(2):e1602.
Tartaglia LA, Gimeno CJ, Storz G, Ames BN. Multidegenerate DNA recognition by the OxyR transcriptional regulator. J Biol Chem. 1992;267:2038–45.
Toledano MB, Kullik I, Trinh F, Baird PT, Schneider TD, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909.
González-Flecha B, Demple B. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem. 1995;270:13681–7.
Charoenlap N, Jiramonai L, Chittrakanwong J, Tunsakul N, Mongkolsuk S, Vattanaviboon P. Inactivation of ahpC renders Stenotrophomonas maltophilia resistant to the disinfectant hydrogen peroxide. Antonie Van Leeuwenhoek. 2019;112:809–14.
Hassett DJ, Alsabbagh E, Parvatiyar K, Howell ML, Wilmott RW, Ochsner UA. A protease-resistant catalase, KatA, released upon cell lysis during stationary phase is essential for aerobic survival of a Pseudomonas aeruginosa oxyR mutant at low cell densities. J Bacteriol. 2000;182:4557–63.
Acknowledgements
Not applicable.
Funding
This study was funded by grant MOST 108–2320-B-010-032-MY3 from the Ministry of Science and Technology of Taiwan, grants V108B-037 and V109C-195 from Taipei Veterans General Hospital, and grant 2019SKHAND007 from the Shin Kong Wu Ho-Su Memorial Hospital, Taipei, Taiwan.
Author information
Authors and Affiliations
Contributions
LHL and YLS conceived the experiment and analyzed data. JYH, CJW, YWH, HHH, and YCT performed the experiments, generated and collected the data. TCY designed the experiment, interpreted the data, and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1 Table S1.
Bacterial strains, plasmids and primers used in this study.
Additional file 2 Fig. S1.
Analysis of putative OxyR binding motifs in the upstream regions of katA2 and ahpC genes.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, LH., Shih, YL., Huang, JY. et al. Protection from hydrogen peroxide stress relies mainly on AhpCF and KatA2 in Stenotrophomonas maltophilia. J Biomed Sci 27, 37 (2020). https://doi.org/10.1186/s12929-020-00631-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12929-020-00631-4