Abstract
Background
The pathogenesis of rheumatoid arthritis (RA) is an immune imbalance, in which various inflammatory immune cells and pro-inflammatory factors are involved. Interleukin-17 (IL-17), a potent pro-inflammatory cytokine, has been found to have increased expression in the joints of patients with RA compared to healthy individuals. However, the causal relationship between the expression level of IL-17 or IL-17 receptor (IL-17R) and RA remained unknown. In this study, two-sample Mendelian randomization (MR) was used to investigate the causal relationship between IL-17 and RA.
Methods
Summary statistics for RA (14,361 RA cases and 43,923 healthy controls) and IL-17 (3,301 samples) were obtained from an available meta-analysis of published genome-wide association studies (GWAS). Relevant single nucleotide polymorphisms (SNPs) were selected by executing quality control steps from the GWAS summary results. Then we used bi-directional two-sample Mendelian randomization (MR) and multi-variable MR (MVMR) analysis to examine evidence of causality. MR and MVMR analyses progressed mainly using inverse variance weighted (IVW), weighted median (WM), and MR-Egger regression methods, which were applied to the genetic instrumental variables (IVs) of IL-17A/IL-17 RA, IL-17C/IL-17 RC, and IL-17D/IL-17RD and RA. For assessing the robustness of the results, we also carried out a sensitivity analysis to assess heterogeneity and pleiotropy, such as MR-Egger, leave-one-out, and MR pleiotropy residual sum and outlier (MR-PRESSO).
Results
Two-sample MR Analysis showed the causal relationship between IL-17A/IL-17RA and RA. The presence of genetically high IL-17A/IL-17RA may increase the risk of RA (IL-17A(OR = 1.095; 95% C.I., 0.990-1.210, p.adj = 0.013), IL-17RA(OR = 1.113, 95%CI = 1.006-1.231, p.adj = 0.006)). However, the results indicated that IL-17C/IL-17RC, and IL-17D/IL-17RD demonstrated no causal impact on RA (IL-17C(OR = 1.007, 95%CI = 0.890-1.139, p.adj = 0.152), IL-17RC(OR = 1.006, 95%CI = 0.904-1.119, p.adj = 0.152), IL-17D(OR = 0.979, 95%CI = 0.843-1.137, p.adj = 0.130), IL-17RD(OR = 0.983, 95%CI = 0.876-1.104, p.adj = 0.129)). Furthermore, MVMR analysis shown that IL-17RA(OR = 1.049, 95% CI: 0.997-1.102, p.adj = 0.014) was associated with increased risk of RA. Sensitivity analysis showed no heterogeneity and pleiotropy, suggesting that the above results were robust and reliable.
Conclusion
The MR analysis provides evidence that IL-17A/IL-17RA are risk factors for RA. This emphasizes the importance of intervention on IL-17A/IL-17RA in patients with RA. Developing drugs that limit IL-17A may reduce the risk of RA.
Similar content being viewed by others
Introduction
Rheumatoid arthritis (RA) is a typical systemic autoimmune disease characterized by chronic joint inflammation and bone destruction [1,2,3]. It comprises symmetric polyarthritis affecting diarthrodial joints and periarticular structures and presenting several systemic manifestations [4]. A variety of inflammatory immune cells and proinflammatory factors are directly involved in the RA process [5,6,7]. Among them, interleukin-17 (IL-17), a potent pro-inflammatory cytokine, has been found to have increased expression in the joints of RA patients compared to healthy individuals. IL-17 is produced by many types of adaptive and innate immune cells [8], which protects the host from fungal and extracellular bacterial infection and plays an essential role in the pathogenesis of RA [9]. However, the causal relationship between the expression levels of various subtypes of IL-17 or IL-17 receptor (IL-17R) and RA remained unclear.
The IL-17 family consists of six structurally related cytokines, namely IL-17A through IL-17F. The IL-17 receptor family comprises five subunits termed Interleukin-17A receptor (IL-17RA) through IL-17RE [10,11,12]. Ligation of IL-17R by IL-17 activates the phosphorylation of Mitogen-Activated Protein Kinases (MAPK) and nuclear factor-κB(NF-κB) pathway to induce pro-inflammatory cytokines in tissue homeostasis [13, 14]. Those pro-inflammatory cytokines boost some sites, such as the synovial tissue, for the development of inflammation, making further efforts to promote the occurrence of RA [15]. Although some literature suggests that IL-17 or IL-17R and RA maybe have a connection, it remains unclear whether the causal relationship between them [9]. So, we researched the relationship of those factors based on predecessors by MR.
Mendelian randomization (MR) selected single nucleotide polymorphism (SNP) as an instrumental variable (IV) to test the causal relationship between exposure and outcome by using genetic markers associated with exposure [9]. The advantages of using MR for causal inference are as follows. MR used the genetic variation index to measure the causality of disease-related risk factors, overcoming the bias caused by confounding or reverse causality inherent in observational studies [16]. MR can also evaluate the robustness of causal effect estimates by testing the existence of pleiotropy [17]. Two-sample MR analysis allows the use of aggregate statistics from genome-wide association studies (GWAS) for MR studies without directly analyzing individual-level data [18]. Based on public GWAS data from a large number of people, we use a two-sample MR analysis to illustrate the impact of IL-17 on RA.
Methods
Study design
In this study, we conducted a two-sample MR to evaluate the causal association between RA and IL-17. Relevant SNPs were selected by executing quality control steps from the GWAS summary results. To maximize the accuracy of results, the elected SNPs as IVs, which need to meet three basic assumptions (Fig. 1): (i)SNP was strongly associated with exposure (correlation hypothesis); (ii)SNP was not associated with confounding factors, meaning that the results were not affected by confounding factors (independence assumption); (iii)SNP was not associated with the outcome or did not directly affect the outcome (exclusivity hypothesis). All original studies acquired ethical review approval and informed consent.
Genetic datasets for IL-17
Genetic prediction of the exposure to the IL-17-related gene GWAS data were retrieved from a recent GWAS of the human plasma proteome results from the INTERVAL study with 3301 healthy individuals of European descent (mean age 43.7 years, 51.1% men). The study performed the SomaLogic method to assess 3,622 plasma proteins [19], covering 10,534,735 genotyped SNPs. We applied the IL-17A/ IL-17 RA, IL-17C/ IL-17 RC, and IL-17D/IL-17RD to explore the causal relationship between IL17 and RA.
Genetic datasets associated with RA
The RA-related outcome data set comes from a previous summary data from GWAS conveyed by Okada et al. [20], including 14,361 cases and 43,923 healthy controls of European descent with a total of 8,747,963 SNPs related to RA. All RA cases fulfilled the 1987 RA diagnosis criteria of the American College of Rheumatology [21] or were diagnosed as RA by a professional rheumatologist.
Selection of IVs
Several quality control steps were taken in our analysis to select qualified IVs closely associated with IL-17. First, to satisfy the correlation hypothesis, we deleted SNPs with a minor allele frequency (MAF) < 1% to enhance the statistical power of genetic variants. Moreover, we removed variants with a physical distance of less than 10,000 kb and r2 < 0.001 to avoid linkage disequilibrium (LD). In the process of selecting independent snps that are closely related to IL-17 and have p-values less than 5e-8, we found that 1 SNP was associated with IL-17A, IL-17RC and IL-17RD, 3 SNPs were connected with IL-17RA, none SNP were associated with IL-17C and IL-17D. In order to satisfy enough eligible IVs for subsequent studies, we broadened the threshold to 1e-5. We also analyzed whether these SNPs were possible confounding factors in PhenoScanner (http://www.phenoscanner.medschl.cam.ac.uk/) to satisfy the independence hypothesis, such as smoking, alcohol consumption, body mass index, and SNPs associated with these potential aspects were excluded. Finally, to guarantee that the effect of a SNP on the exposure and the effect of that SNP on the outcome must each correspond to the same allele, we harmonized the exposure and outcome datasets by correct the strand for non-palindromic SNPs and drop all palindromic SNPs. We utilized these carefully chosen SNPs as the final genetic IVs for the subsequent MR analysis.
The F statistic is used to test the correlation hypothesis that SNP was strongly associated with exposure, calculated using the formula: \(\mathrm{F}=\frac{{\mathrm{R}}^{2}\left(\mathrm{n}-\mathrm{k}-1\right)}{\left(1-{\mathrm{R}}^{2}\right)\mathrm{k}}\). R2 denotes the variance of exposure explained by IVs, n is the sample size and k is the number of instrumental variables [22]. For Mendelian randomization, the F statistic is an indicator of the strength of the IVs, with values over 10 reflecting strong instruments. If the F statistic less than ten, IVs were considered weak instruments and would be excluded for MR analysis [23].
Statistical analysis
We utilized these MR methods from TwoSampleMR packages in R software (version 4.1.0) to estimate the causal effects of the cytokines and cytokine receptors (A, C, D) of IL-17 on RA. We used inverse variance weighted (IVW), weighted median (WM), and MR-Egger regression methods to assess the causal influence of the exposure on the outcome [24,25,26]. IVW is the primary analysis method that can balance pleiotropy. MR-egger and weighted median enhance the estimation of IVW, although less efficiently [27].
Sensitivity analysis
To assess the robustness of these results and prevent potential pleiotropy and heterogeneity, a series of sensitivity analyses, including MR-Egger intercept tests, MR-pleiotropy residual sum and outlier (MR-PRESSO) global test, and Cochran's Q test for heterogeneity, Funnel pot test, leave-one-out analyses. A P value of < 0.05 in the Cochran Q test was considered statistically significant. MR-egger was used to evaluate the potential horizontal pleiotropy. The intercept term of MR-Egger regression was used to assess whether horizontal pleiotropy affected the results of MR analysis, where P < 0.05 indicated horizontal pleiotropy [28]. By visual inspection of funnel plots, asymmetries indicate horizontal pleiotropy. We performed a Leave-one-out analysis to determine whether any single SNP drove the causal estimates. We repeated the IVW analysis by discarding each exposure-related SNP [24]. In the MR-PRESSO test, SNPs associated with heterogeneity were eliminated to reduce outliers in the estimation of causal effects [29].
Bonferroni correction method was used to account for multiple testing in this study. Associations with adjust p value (p.adj) < 0.05 were regarded as significant associations. Since this MR study was conducted using publicly available GWAS summary data, ethical approval and informed consent from all subjects could be found in the original publications.
Result
IVs for MR
The selection process of IL-17RA-related SNPs is detailed below, and other exposure-related SNPs are shown in Table 1. A total of 25 SNPs associated with IL-17RA were identified after the genome-wide significance threshold (p < 1 × 10–5) and clumping (r2 < 0.001). Among them, 6 SNPs were correlated with outcome RA after the harmonizing process, and none of them was an outlier variant detected by the MR-PRESSO test. We checked in PhenoScanner V2 whether the previously retained SNPs were connected with confounding factors, such as smoking, drinking, and body mass index (BMI), and there were no abnormal SNPs. Finally, 6 SNPs were chosen as instrumental variables for IL-17RA in the present study.
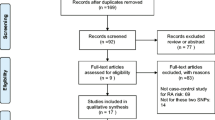
After the same quality control steps, 12 independent SNPs were related to IL-17A, 9 independent SNPs were associated with IL-17C, 13 independent SNPs were connected with IL-17RC, 10 independent SNPs were related with IL-17D, 11 independent SNPs were correlated with IL-17RD. Among the SNPs connected with IL-17RD, rs6776722 (associated with alcohol) and rs400824 (associated with BMI) were excluded. The selection process of IVs for MR analysis is shown in Fig. 2 and these SNPs are shown in detail in Supplementary Table 1.
Mendelian randomization results
We evaluated the causal effect of IL-17A/IL-17RA, IL-17C/IL-17RC, and IL-17D/IL-17RD on RA using two-sample MR methods (Table 1). After Bonferroni-correction, according to the IVW and weighted-median analysis, the presence of genetically high IL-17A/IL-17RA were associated with increased risk of RA (IL-17A(OR = 1.095; 95% C.I., 0.990–1.210, p.adj = 0.013), IL-17RA(OR = 1.113, 95%CI = 1.006–1.231, p.adj = 0.006)).. In addition, IVW analyses showed that IL-17C/IL-17RC, and IL-17D/IL-17RD demonstrated no causal impact on RA (IL-17C(OR = 1.007, 95%CI = 0.890–1.139, p.adj = 0.152), IL-17RC(OR = 1.006, 95%CI = 0.904–1.119, p.adj = 0.152), IL-17D(OR = 0.979, 95%CI = 0.843–1.137, p.adj = 0.130), IL-17RD(OR = 0.983, 95%CI = 0.876–1.104, p.adj = 0.129)), and these results were confirmed by the weighted-median and MR-Egger analyses. Forest plots and scatter plots of the association between IL-17A/IL-17RA, IL-17C/IL-17RC, and IL-17D/IL-17RD and RA are shown in Fig. 3 and Fig. 4.
Sensitivity analysis
Regarding heterogeneity and sensitivity, Cochran’s Q-test (PQ > 0.05) and MR-Egger regression indicated no heterogeneity in the causal effect between IL-17A/ IL-17 RA, IL-17C/ IL-17 RC, IL-17D/IL-17RD, and RA. The funnel plot displayed a symmetric pattern of effect size variation around the point estimate (Fig. 5). In the leave-one-out analyses, we found that the risk estimates of IL-17 levels and risk of RA kept consistent substantially after excluding one SNP at each time, which means IL-17-RA association was not driven by any individual SNP exclusion, suggesting that SNPs without potential influence causal relationship and the conclusion was stable and reliable (Fig. 6). As for pleiotropic analysis, the pooled causal effects of the MR-Egger regression analysis were consistent with the IVW results, indicating that all variables were effective for RA. MR-PRESSO indicated no presence of potential pleiotropy in our tests.
Multivariable MR analysis
In model with mutual adjustment for IL17A, IL17C, IL17D, IL17RA, IL17RC and IL17RD, the association between IL17RA and risk of RA was significant (nSNP = 52, OR = 1.049, 95% CI: 0.997–1.102, p.adj = 0.014, Table2). This finding again suggested the IL17RA is one of risk factor for RA.
Discussion
This study used MR analyses firstly to investigate potential causal links between a set of IL-17 family and RA, which found that IL-17A/IL-17RA maybe contribute to the higher risk of RA. However, there were no causal effects of IL-17C/ IL-17RC, and IL-17D/ IL-17RD on RA according to the MR results. These findings would contribute greatly to research on the mechanism and treatment of RA.
Studies in the 1990s showed that compared to healthy individuals, IL-17 expression had increased in the joint of RA patients [30, 31]. And high levels of serum IL-17 A have been reported in RA [32]. In addition, IL-17RA were over-expressed in RA peripheral blood, and their expression was detected locally in RA synovium [33]. IL-17A is involved in inflammation and defense against infection by inducing fibroblasts, endothelial cells, and epithelial cells to express pro-inflammatory cytokines (TNF, IL-1, IL-6, G-CSF, and GM-CSF), chemokines (CXCL1, CXCL5, IL-8, CCL2, and CCL7), and antimicrobial peptides (defensin and S100 protein) and matrix metalloproteinases (MMP1, MMP3, and MMP13) [11, 34, 35]. IL-17A promotes granulocyte formation mediated by stem cell factor and granulocyte colony-stimulating factor and recruits neutrophils to inflammatory sites [31, 32, 36]. IL-17A induces keratinocytes to express intercellular adhesion molecule-1 (ICAM-1) and chondrocytes to express iNOS and cyclooxygenase-2 [37, 38]. These may explain why IL-17A/IL-17RA is a risk factor for RA. However, we found no direct causal relationship between IL-17C/IL-17RC, and IL-17D/IL-17RD and RA. They may participate in the occurrence and development of RA through other risk factors or pathways.
Drugs that restrict IL-17A may reduce the risk of RA. Currently, monoclonal antibodies (Mabs) targeting IL-17 or IL-17R include Ixekizumab, Secukinumab, Brodalumab, and bimekizumab. Ixekizumab (Taltz®, LY2439821) is a humanized IgG4 monoclonal antibody [39]. Secukinumab (AIN457, Cosentyx) is a recombinant human IgG1/kappa monoclonal antibody [40, 41]. Brodalumab (AMG 2) is a fully human IgG17 monoclonal antibody [42]. Bimekizumab (496.g3, formerly UCB4940) is a humanized monoclonal antibody with a high affinity for IL-17A and IL-17F [43]. They all target IL-17A and inhibit its binding and interaction with its receptors, such as blocking IL-17A, IL-17F, and IL-17-A/F heterodimer signaling and IL-17E signaling in IL-17RA/RC complexes. The secretion of protein kinases, pro-inflammatory cytokines, and chemokines is then inhibited by targeting cells with downstream effects on cellular elements [8].
There are several advantages to our MR analysis. Compared with randomized controlled trials, when selecting appropriate tool variables, this method reduced the interference of confounding factors to a greater extent. In our study, we excluded the influence of horizontal pleiotropy, further indicating the study's reliability. Additionally, we analyzed all IL-17 and IL17RA one by one, which highly improved universality and expansibility. Even so, there are several limitations of the study. For example, the summary GWAS data we mainly used refers to individuals of European ancestry, so it should be considered accurate when the results are applied to other races. GWAS are able to identify common variants with moderate to small effect sizes, but they may not be sensitive to identifying genes with disease-causing mutations. In addition, we could not further explore the MR effect estimates between IL-17 levels and disease severity due to the lack of relevant data. Moreover, the possible causal relationship between IL-17RA and RA found in the current study will require experimental verifications in a laboratory animal model(s) with careful controlled randomized study design.
Conclusion
In conclusion, the MR analysis supported a causal relationship between genetically predicted IL-17RA and RA. This study provided strong evidence that IL-17RA is a risk factor for RA. What's more, it will promote studies on reducing the risk of RA by developing drugs that limit IL-17A.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [Supplementary Table 1].
Abbreviations
- RA:
-
Rheumatoid arthritis
- IL-17:
-
Interleukin-17
- IL-17RA:
-
Interleukin-17A receptor
- MAPK:
-
Mitogen-Activated Protein Kinases
- NF-κB:
-
Nuclear factor-κB
- MR:
-
Mendelian randomization
- SNP:
-
Single nucleotide polymorphism
- IV:
-
Instrumental variable
- GWAS:
-
Genome-wide association study
- MAF:
-
Minor allele frequency
- LD:
-
Linkage disequilibrium
- IVW:
-
Inverse variance weighted
- WM:
-
Weighted median
- MR-PRESSO:
-
MR-pleiotropy residual sum and outlier
- BMI:
-
Body mass index
- Mabs:
-
Monoclonal antibodies
- OR:
-
Odds ratio
- CI:
-
Confidence interval
References
Hsu YH, Chang MS. IL-20 in rheumatoid arthritis. Drug Discov Today. 2017;22(6):960–4.
Giannini D, Antonucci M, Petrelli F, Bilia S, Alunno A, Puxeddu I. One year in review 2020: pathogenesis of rheumatoid arthritis. Clin Exp Rheumatol. 2020;38(3):387–97.
Viatte S, Barton A. Genetics of rheumatoid arthritis susceptibility, severity, and treatment response. Semin Immunopathol. 2017;39(4):395–408.
Macedo RB, Kakehasi AM, Melo de Andrade MV. IL33 in rheumatoid arthritis: potential contribution to pathogenesis. Rev Bras Reumatol Engl Ed. 2016;56(5):451–7.
Wasserman AM. Diagnosis and management of rheumatoid arthritis. Am Fam Physician. 2011;84(11):1245–52.
Huang J, Fu X, Chen X, Li Z, Huang Y, Liang C. Promising Therapeutic Targets for Treatment of Rheumatoid Arthritis. Front Immunol. 2021;12: 686155.
Kondo N, Kuroda T, Kobayashi D. Cytokine Networks in the Pathogenesis of Rheumatoid Arthritis. Int J Mol Sci. 2021;22(20):10922.
Berry SPD, Dossou C, Kashif A, Sharifinejad N, Azizi G, Hamedifar H, et al. The role of IL-17 and anti-IL-17 agents in the immunopathogenesis and management of autoimmune and inflammatory diseases. Int Immunopharmacol. 2022;102: 108402.
Omidian Z, Ahmed R, Giwa A, Donner T, Hamad ARA. IL-17 and limits of success. Cell Immunol. 2019;339:33–40.
Amatya N, Garg AV, Gaffen SL. IL-17 Signaling: The Yin and the Yang. Trends Immunol. 2017;38(5):310–22.
Kirkham BW, Kavanaugh A, Reich K. Interleukin-17A: a unique pathway in immune-mediated diseases: psoriasis, psoriatic arthritis and rheumatoid arthritis. Immunology. 2014;141(2):133–42.
Wang J, He L, Li W, Lv S. A Role of IL-17 in Rheumatoid Arthritis Patients Complicated With Atherosclerosis. Front Pharmacol. 2022;13: 828933.
Chai BY, Yip WK, Dusa N, Mohtarrudin N, Seow HF. Loss of Interleukin-17RA Expression is Associated with Tumour Progression in Colorectal Carcinoma. Pathol Oncol Res. 2020;26(4):2291–8.
Tournadre A, Miossec P. Interleukin-17 in inflammatory myopathies. Curr Rheumatol Rep. 2012;14(3):252–6.
Kuwabara T, Ishikawa F, Kondo M, Kakiuchi T. The Role of IL-17 and Related Cytokines in Inflammatory Autoimmune Diseases. Mediators Inflamm. 2017;2017:3908061.
Cui Z, Hou G, Meng X, Feng H, He B, Tian Y. Bidirectional Causal Associations Between Inflammatory Bowel Disease and Ankylosing Spondylitis: A Two-Sample Mendelian Randomization Analysis. Front Genet. 2020;11: 587876.
Qu Z, Huang J, Yang F, Hong J, Wang W, Yan S. Sex hormone-binding globulin and arthritis: a Mendelian randomization study. Arthritis Res Ther. 2020;22(1):118.
Chen X, Kong J, Pan J, Huang K, Zhou W, Diao X, et al. Kidney damage causally affects the brain cortical structure: A Mendelian randomization study. EBioMedicine. 2021;72: 103592.
Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558(7708):73–9.
Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–81.
Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31(3):315–24.
Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–64.
Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol. 2013;178(7):1177–84.
Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377–89.
Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40(4):304–14.
Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7. https://doi.org/10.7554/eLife.34408.
Ong JS, MacGregor S. Implementing MR-PRESSO and GCTA-GSMR for pleiotropy assessment in Mendelian randomization studies from a practitioner’s perspective. Genet Epidemiol. 2019;43(6):609–16.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34.
Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8.
Schinocca C, Rizzo C, Fasano S, Grasso G, La Barbera L, Ciccia F, et al. Role of the IL-23/IL-17 Pathway in Rheumatic Diseases: An Overview. Front Immunol. 2021;12: 637829.
Iwakura Y, Nakae S, Saijo S, Ishigame H. The roles of IL-17A in inflammatory immune responses and host defense against pathogens. Immunol Rev. 2008;226:57–79.
Ruiz de Morales JMG, Puig L, Daudén E, Cañete JD, Pablos JL, Martín AO, et al. Critical role of interleukin (IL)-17 in inflammatory and immune disorders: An updated review of the evidence focusing in controversies. Autoimmun Rev. 2020;19(1):102429.
Zrioual S, Toh ML, Tournadre A, Zhou Y, Cazalis MA, Pachot A, et al. IL-17RA and IL-17RC receptors are essential for IL-17A-induced ELR+ CXC chemokine expression in synoviocytes and are overexpressed in rheumatoid blood. J Immunol. 2008;180(1):655–63.
Li G, Zhang Y, Qian Y, Zhang H, Guo S, Sunagawa M, et al. Interleukin-17A promotes rheumatoid arthritis synoviocytes migration and invasion under hypoxia by increasing MMP2 and MMP9 expression through NF-κB/HIF-1α pathway. Mol Immunol. 2013;53(3):227–36.
Koper-Lenkiewicz OM, Sutkowska K, Wawrusiewicz-Kurylonek N, Kowalewska E, Matowicka-Karna J. Proinflammatory Cytokines (IL-1, -6, -8, -15, -17, -18, -23, TNF-α) Single Nucleotide Polymorphisms in Rheumatoid Arthritis-A Literature Review. Int J Mol Sci. 2022;23(4):365.
Taams LS. Interleukin-17 in rheumatoid arthritis: Trials and tribulations. J Exp Med. 2020;217(3):e20192048.
Chen S, Blijdorp IC, van Mens LJJ, Bowcutt R, Latuhihin TE, van de Sande MGH, et al. Interleukin 17A and IL-17F Expression and Functional Responses in Rheumatoid Arthritis and Peripheral Spondyloarthritis. J Rheumatol. 2020;47(11):1606–13.
Iwakura Y, Ishigame H, Saijo S, Nakae S. Functional specialization of interleukin-17 family members. Immunity. 2011;34(2):149–62.
Persson EC, Engström C, Thilander B. The effect of thyroxine on craniofacial morphology in the growing rat. Part I: A longitudinal cephalometric analysis. Eur J Orthod. 1989;11(1):59–66.
Blauvelt A. Safety of secukinumab in the treatment of psoriasis. Expert Opin Drug Saf. 2016;15(10):1413–20.
Koenders MI, van den Berg WB. Secukinumab for rheumatology: development and its potential place in therapy. Drug Des Devel Ther. 2016;10:2069–80.
Lebwohl M, Strober B, Menter A, Gordon K, Weglowska J, Puig L, et al. Phase 3 Studies Comparing Brodalumab with Ustekinumab in Psoriasis. N Engl J Med. 2015;373(14):1318–28.
Adams R, Maroof A, Baker T, Lawson ADG, Oliver R, Paveley R, et al. Bimekizumab, a Novel Humanized IgG1 Antibody That Neutralizes Both IL-17A and IL-17F. Front Immunol. 2020;11:1894.
Acknowledgements
Thank Okada et al., Sun et al. and the genome-wide association study consortia who made their summary statistics publicly available for this study without whom this effort would not be possible, and thanked the staff and participants of the study for their important contributions.
Funding
No funding.
Author information
Authors and Affiliations
Contributions
Dr Rong Zhao and Yi-wen Zhang contributed equally to this paper and share first authorship. Study design, data analysis, and manuscript writing: Rong Zhao, Yiwen Zhang and Jiayuan Yao. Review and editing, supervision, and funding acquisition: Rong Zhao, Shengxiao Zhang, Jun Qiao and Shan Song. All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary table 1.
Summary of the 61 SNPs associated with IL-17.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhao, R., Zhang, Yw., Yao, Jy. et al. Genetic association between interleukin-17 and susceptibility to rheumatoid arthritis. BMC Med Genomics 16, 277 (2023). https://doi.org/10.1186/s12920-023-01713-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-023-01713-6