Abstract
Background
Congenital heart disease (CHD) is one of the most common birth defects. Copy number variations (CNVs) have been proved to be important genetic factors that contribute to CHD. Here we screened genome-wide CNVs in Chinese children with complete atrioventricular canal (CAVC) and single ventricle (SV), since there were scarce researches dedicated to these two types of CHD.
Methods
We screened CNVs in 262 sporadic CAVC cases and 259 sporadic SV cases respectively, using a customized SNP array. The detected CNVs were annotated and filtered using available databases.
Results
Among 262 CAVC patients, we identified 6 potentially-causative CNVs in 43 individuals (16.41%, 43/262), including 2 syndrome-related CNVs (7q11.23 and 8q24.3 deletion). Surprisingly, 90.70% CAVC patients with detected CNVs (39/43) were found to carry duplications of 21q11.2–21q22.3, which were recognized as trisomy 21 (Down syndrome, DS). In CAVC with DS patients, the female to male ratio was 1.6:1.0 (24:15), and the rate of pulmonary hypertension (PH) was 41.03% (16/39). Additionally, 6 potentially-causative CNVs were identified in the SV patients (2.32%, 6/259), and none of them was trisomy 21. Most CNVs identified in our cohort were classified as rare (< 1%), occurring just once among CAVC or SV individuals except the 21q11.2–21q22.3 duplication (14.89%) in CAVC cohort.
Conclusions
Our study identified 12 potentially-causative CNVs in 262 CAVC and 259 SV patients, representing the largest cohort of these two CHD types in Chinese population. The results provided strong correlation between CAVC and DS, which also showed sex difference and high incidence of PH. The presence of potentially-causative CNVs suggests the etiology of complex CHD is incredibly diverse, and CHD candidate genes remain to be discovered.
Similar content being viewed by others
Background
Congenital heart disease (CHD) is the most common birth defect with an incidence of 1–1.2% in live births [1, 2]. Due to disrupted early-stage development, CHD consists of many structural malformations of the cardiovascular system, ranging from simple lesions such as atrial septal defects (ASD) and ventricular septal defects (VSD), to complex lesions such as tetralogy of Fallot (ToF), complete atrioventricular canal (CAVC) and single ventricle (SV). Although clinical treatment have significantly improved, complex CHD still remains to be a leading cause of newborn-related mortality [3].
Consistent with the complexity of early heart development, the etiology of CHD is multifactorial. To date, only about 20–30% of CHD cases could be attributed to genetic or environmental causes based on available technologies [4,5,6]. The incidence of some specific CHD types has been revealed with sex or race biases [7, 8]. The recurrence risk of CHD in the offspring of an affected parent, as well as in the siblings of a CHD child, has been reported to be higher than the general population [9, 10]. The evidences emphasize that genetics plays an important role in the pathogenesis of CHD [11].
Both small and large genetic variations could contribute to CHD [12]. Small insertions and deletions (INDELs), ranging from 1 bp to 10 kb in length [13], are typically detected by sequencing technologies [14]. Putative deleterious small variants in single genes could cause both syndromic and isolated CHD. For instance, Noonan syndrome, a common genetic disorder, is mostly caused by mutations in PTPN11 gene. Pulmonary valve stenosis and CAVC represent relatively common features in Noonan syndrome [15, 16]. Heterotaxy syndrome, which comprises a class of congenital disorders resulting from malformations in left–right body patterning, has been reported to be associated with mutations in NODAL, ACVR2B, LEFTY2, GDF1, ZIC3, CRELD1 and NKX2.5 [17]. Majority patients with heterotaxy syndrome have serious CHD including SV [17, 18]. In addition to syndromic CHD, an increasing number of genes have been identified in individuals with isolated CHD [11]. Whole exome sequencing and whole genome sequencing are able to effectively identify small variants associated with CHD.
Large genetic variants, including aneuploidies, chromosomal rearrangements and copy number variations (CNVs), are also important genetic causes of CHD. CNVs can range in size from single genes to large contiguous deletions or duplications of millions of base pairs [19, 20]. Pathogenic CNVs tend to be large, de novo and disrupting coding regions [20]. Although recent advances in next generation sequencing showed their potential in CNVs detection, chromosome microarray, either array comparative genomic hybridization or single nucleotide polymorphism array, is still the gold standard for CNV detection and validation [21].
Nowadays, investigation of genes in overlapping CNV regions can probably identify relevant genes or refined intervals for certain genetic diseases [22,23,24]. Considering the heterogeneity of CHD etiology, a large number of CNVs associated with CHD have been identified over the past decades, especially the conotruncal anomalies including TOF, TGA and pulmonary atresia (PA)/VSD [25,26,27]. In our study, genome-wide CNVs in Chinese children with CAVC and SV were screened since there were scarce researches dedicated to these two types of CHD.
Methods
Study subjects
We obtained a cohort of 528 children diagnosed as CAVC (n = 264) or SV (n = 264) by echocardiography from the Shanghai Children’s Medical Center between November 2010 and August 2019. The patients had an average age at 8.77 ± 2.77 (mean ± SD) years. The phenotypic details of this cohort were summarized in Additional file 1: Table S1. The Ethics Committee of the Shanghai Children’s Medical Center reviewed and approved this study (SCMCIRB-K2017009).
DNA extraction
Genomic DNA was isolated from peripheral blood samples of all patients using Gentra Puregene Blood Kit (QIAGEN, Hilden, Germany) according to manufacturers’ instructions. NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) was used to check the quantity and quality of the DNA samples. Only samples with OD260/OD280 ratios between 1.8 and 2.0, and OD260/OD230 ratios > 1.5 were selected for further investigation.
Microarrays
Microarrays were designed based on the Affymetrix Arrays platforms (Thermo Fisher Scientific, Waltham, MA), namely the CytoScan 750 K arrays. We deleted probes with high population frequency and added probes particularly designed for sites marked with two stars in Clinvar as well as pathogenic variants in HGMD. In the meantime, design of probes was also based on clinical data of high-morbidity diseases in newborns, which was applied to screen for CHD, especially CHD patients accompanied by extra-cardiac anomalies. Genomic DNA samples were amplified, fragmented and stringently hybridized onto arrays according to manufacturers’ instructions. Microarrays were automatedly processed by GeneTitan Multi-Channel instruments together with Affymetrix Command Console (AGCC) Software for instruments control and production of probe cell intensity data (CEL file).
Data analysis
Microarray data processing was implemented using the Affymetrix Chromosome Analysis Suite v2.0 (ChAS) Software, and CNVs were called based on human assembly GRCh38 (hg38). There were 521 patients including 262 CAVC and 259 SV passed the QC tests finally.
Only CNV calls larger than 200 kb and with at least 50 probes for deletion and duplication were considered for further analysis. The detected CNVs calls were identified by public databases and websites: Database of Genomic Variants (DGV, http://dgv.tcag.ca/dgv/app/home), Online Mendelian Inheritance in Man (OMIM, https://omim.org/), UCSC Genome Browser (https://genome.ucsc.edu/), the Clinical Genome Resource (ClinGen, https://www.clinicalgenome.org/), Database of Chromosomal Imbalance, PubMed (https://www.ncbi.nlm.nih.gov/pubmed/), Phenotype of Humans using Ensemble Resources (DECIPHER, http://decipher.sanger.ac.uk/), SCAN (http://www.scandb.org/newinterface/about.html), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
In this study, the detected CNVs were classified according to the following criteria: (1) The ones having ≥ 70% overlap with CNVs reported in DGV were categorized into non-causative CNVs. And the rest CNVs were identified as potentially-causative CNVs. The frequencies for non-causative CNVs and potentially-causative CNVs were calculated based on DGV and DECIPHER database, respectively. (2) In our cohort of 262 CAVC and 259 SV patients, the CNVs had a frequency less than 1% were defined as rare CNVs, and the others (≥ 1% in our dataset) were identified as common CNVs. (3) Novel CNVs were those have not been previously reported in the literature or available public database.
Results
Clinical characteristics of the 528 patients
In 264 CAVC patients, the sex ratio (Males to Females) was 0.98 (131:133) and the average age was 8.80 ± 3.76 (mean ± SD) years. In 264 SV patients, the sex ratio was 1.34 (151:113) and the average age was 7.04 ± 3.57 years. The results were summarized in Table 1.
CNV detection in CHD cases
In this study, seven samples failed the QC criteria (CAVC209, CAVC211, SV134, SV145, SV177, SV181, SV254). Among the rest 521 samples (262 CAVC and 259 SV) who passed the QC tests, a total of 3465 CNVs were detected with a median size of 922.3 kb (max 23.9 Mb, min 51.5 kb). Large CNVs with size greater than 200 kb were selected for further analysis.
Nine large CNVs (according to the filtering criteria) were identified in 44 CAVC cases (16.79%, 44/262), which consists of 4 duplication CNVs involved 406 genes and 5 deletion CNVs affecting 452 genes (see Table 2). We also identified 18 large CNVs in 16 SV cases (6.06%, 16/264), including 13 duplications and 5 deletions (see Table 3).
There were 15 non-causative CNVs identified, including 3 in CAVC samples (Yp11.2, 17q21.2-17q21.31, Yq11.221–11.222) and 12 in SV patients (14q23.1, Xq28, 22q13.1, 2q35, 2q13, 2q14.2, 22q13.1, 7p14.3, 5q13.2, 5q23.2-5q23.3, 17q12, Xp22.2). The rest CNVs were identified as potentially-causative CNVs. Most CNVs identified in our study were rare (< 1%), occurring just once among the CAVC (0.38%, 1/262) or SV samples (0.39%, 1/259) except the 21q11.2-21q22.3 duplication in CAVC cohort (14.89%, 39/262).
Potentially-causative CNVs in CAVC cases
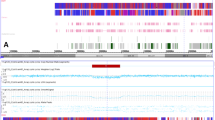
In 262 CAVC patients, 6 potentially-causative CNVs were identified in 43 cases (16.41%, 43/262) (see Table 2). Surprisingly, 90.70% CAVC patients with these CNVs (39/43) were found to carry duplication of 21q11.2–21q22.3 which was considered as a common CNV (14.89%, 39/262) in our cohort. Among them, there was one patient (CAVC274) simultaneously had a ~ 0.8 Mb duplication at 3q12.1–3q12.2 encompassing COL8A1, HP09053, FILIP1L, CMSS1 and TBC1D23. For the deletion CNVs, we had identified isolated deletions of 7q11.23 (CAVC162), 8q21.13-8q21.2 (CAVC102) and 8q24.3 (CAVC145) separately.
Additionally, we also consulted the DECIPHER and ISCA databases for evidences of clinical relevance. Duplication of 9p24.3-9p13.11 (CAVC207) has been reported to associate with ToF/TGA/coarctation of the aorta (CoA) phenotype.
Potentially-causative CNVs in SV cases
Totally, 6 cases with potentially-causative CNVs were identified in 259 SV patients (2.32%, 6/259) (see Table 3), and none of them were identified as trisomy 21. Except deletion of 8q21.13–8q21.3, the rest were all duplication CNVs (5q34-5q35.1, Xq22.1, 1q42.12-1q42.13, 2p13.2, 7p14.1). These CNVs have been reported to associate with VSD, ToF and CoA in DECIPHER and ISCA.
Additionally, we noticed that two patients were simultaneously identified with two CNVs. One patient (SV143) was identified with one non-causative CNV (2q14.2, dup) and one potentially-causative CNV (5q34–5q35.1, dup), and the other patient (SV261) owned one non-causative CNV (Xp22.2, dup) as well as one potentially-causative CNV (7p14.1, dup).
Discussion
CAVC, accounts for ~ 4% of CHD, is a complex cardiac malformation characterized by a variable deficiency of the atrioventricular area in the developing heart [28, 29]. SV, one of the most common forms of severe CHD, comprises a spectrum of congenital cardiac malformations defined by severe underdevelopment of one ventricle [30].
In 262 CAVC patients reported here, 14.89% (39/262) carried the duplication of 21q11.2-21q22.3, which could be diagnosed as trisomy 21, namely Down Syndrome (DS). A striking association of CAVC with DS was found in this study. All DS patients had the same ~ 3.3 Mb duplication at 21q11.2-21q22.3, and a systematic reanalysis indicated that 21q22.13 was the minimal critical region to the DS phenotype [31]. Additionally, another study detected 57.6% cardiac malformations in 500 patients with DS, and it also suggested CAVC (35.1%) was the most frequent heart anomaly [32]. It is putative that CAVC is the most frequent type of CHD in DS patients, and our study also provide strong evidence for this correlation in Chinese population. Additionally, CAVC also referred to as complete atrioventricular septal defect, and it has been reported that AVSD (atrioventricular septal defects) are more common in the female of DS patients [33]. In our study, the female/male ratio of CAVC with DS patients was 1.60 (24:15), which suggest that potential sex differences existed in the prevalence of CAVC in DS patients. Besides, we also noticed that rates of pulmonary hypertension (PH) in DS patients with CAVC was 41% (16/39), which was higher than previous report (28%, 364/1242) [34]. In fact, it was well known that PH is common in children with DS, and our study intensely proved this correlation.
Nowadays, several genes located in the “CHD critical region” on chromosome 21 have been proved to be associated with CAVC, including DSCAM, COL6A1, COL6A2, and DSCR1 [35]. However, there were three DS patients simultaneously had another CNV located at different chromosome in our cohort, and one of the CNVs (3q12.1–3q12.2 dup) has been reported to associate with VSD in Decipher database. Additionally, several DS patients showed not only CAVC (4/27), but also other cardiac anomalies, such as ToF, ASD, patent foramen oval (PFO) and patent ductus arteriosus (PDA). Although the above-mentioned genes can explain partial cardiac phenotypes in DS patients, the genetic causes still were difficult to clarify especially when DS probands accompanied with multiple CNVs and diverse CHD phenotypes.
In our study, two potentially-causative CNVs had been identified as main causes of certain syndromes with heart anomalies. The microdeletion on 7q11.23 caused Williams-Beuren Syndrome (WBS; OMIM 194050), which is a multisystemic developmental disorder mostly accompanied with CHD [36, 37]. More than 90% of WBS patients have the ~ 1.55 Mb pair deletion extending from FKBP6 to GTF2I, and it has been widely accepted that the deletion or mutation of an elastin (ELN) allele is a major cause of WBS [38]. One patient in this study (CAVC162) had a ~ 1.52 Mb deletion at 7q11.23 extending from NCF1B to GTF2I, encompassing the ELN gene. The other microdeletion on 8q24.3 have been recognized as associated with Verheij syndrome (OMIM 615583), which is characterized by growth retardation, developmental delay (DD), microcephaly, vertebral anomalies, dysmorphic features, cardiac and renal defects [39]. Poly(U) Binding Splicing Factor 60 (PUF60) were suggested as the main cause for heart defects in the syndrome, since knockdown of Puf60 alone resulted in cardiac structural defects [40]. The patient (CAVC145) reported here had a ~ 2.5 Mb deletion of 8q24.3, representing with growth retardation and heart anomalies.
Among the rest CNVs identified in the CAVC patients, CNVs located at 8q21.13–8q21.2, 9p24.3–9p13.1 and 3q12.1–3q12.2 have been seldom reported. The detected potentially-causative deletion CNV, 8q21.13–8q21.2, encompasses Zinc Finger and BTB Domain Containing 10 (ZBTB10), which has been known as a CHD gene. ZBTB10 encodes a telomere-associated protein [41]. Lately, a GWAS involving 4,000 unrelated Caucasian patients diagnosed with CHD indicated that ZBTB10 was associated with TGA, since two highly significant SNPs (rs148563140 and rs143638934) closely located to this gene [42]. Furthermore, they suggested strong cell-type specificity in murine cardiac development for Zbtb10. Except the known CHD gene ZBTB10, this CNV region in the patient (CAVC102) also included STMN2 related to abnormality of the cardiovascular system. STMN2 encodes a member of the stathmin family of phosphoproteins, functioning in microtubule dynamics and signal transduction [43]. Compared with controls, methylation of STMN2 significantly increased (FDR p value = 4.27 × 10–51) in VSD cases [44]. Besides, it has been shown that Stmn2 expresses in atrioventricular node, endocardium and outflow tract in mouse according to the LifeMap Discovery database. For the remaining two duplication CNVs, one of them (9p24.3–9p13.1, CAVC207) has been reported as VSD or TOF in DECIPHER. In this region, only Rfx3 gene was in “ventricular septal defect” derived from the MGI (mouse genome informatics) database. For the duplicated region of 3q12.1–3q12.2, a report had shown a VSD patient had a ~ 116 kb duplication of this region, and TBC1D23 has been identified as the major candidate gene [25]. In our study, the patient (CAVC274) had a ~ 0.8 Mb duplication at 3q12.1–3q12.2, encompassing this CHD candidate gene TBC1D23.
There were 6 potentially-causative CNVs (8q21.13–8q21.3, 1q42.12–1q42.13, Xq22.1, 5q34–5q35.1, 2p13.2, 7p14.1) in 259 SV patients. The deleted region of 8q21.13-8q21.3 (SV007) overlapped with the above-mentioned CNVs in the CAVC patient (CAVC102). Furthermore, we noticed that duplication of 1q42.12–1q42.13 (SV163) included 2 known CHD risk genes, LEFTY1 and LEFTY2 [45]. Especially, It has been reported that the SNP rs2295418 in the LEFTY2 gene is associated with CHD in Chinese Han populations [46]. For other detected CNVs, only several genes were included in the deleted or duplicated regions, namely Xq22.1 (SV147; PCDH19, TNMD), 2p13.1 (SV195; DYSF, CYP26B1, EXOC6B) and 7p14.1 (SV176, INHBA and SUGCT). There were no related reports between CHD and these detected CNVs, so we focused on three genes (PCDH19, CYP26B1 and INHBA) with a significantly higher pLI score, which reflects the intolerance to the loss of function mutations. PCDH19 (Protocadherin 19) is a member of the delta-2 protocadherin subclass of the cadherin superfamily. CYP26B1 (cytochrome P450 family 26 subfamily B member 1) involves in limiting retinoic acid (RA) levels within vertebrate embryos, which facilitate RA degradation [47]. It has been well-known that RA is important for the development of the heart. INHBA (Inhibin Subunit Beta A) encodes a member of the TGF-beta (transforming growth factor-beta) superfamily of proteins, and it has been shown as a candidate gene for cardiac development [48].
As for the proband (SV143) with a ~ 1.6 Mb duplication at 5q34-5q35.1, we found that this region encompasses 2 genes (SLIT3 and TENM2) related to septal defects of heart. SLIT3 (Slit Guidance Ligand 3) expressed in cardiomyocyte-like progenitor cells [49], and membranous ventricular septum defects as well as atrioventricular and aortic valve abnormalities are exhibited in SLIT3-mutant mice [50]. Recently, SLIT3 variants in humans has shown association with CHD involving in ToF and septal and outflow tract defects [51]. TENM2 (Teneurin Transmembrane Protein 2) expresses abundantly in human fetal heart. Moreover, patients with loss of TENM2 presented ASD in Decipher database, gain of TENM2 didn’t show any phenotype of CHD yet.
Structural genetic changes, especially copy number variants, represent a major source of genetic variation contributing to CHD patients. In recent years, a large number of CNVs associated with CHD have been identified [25,26,27]. Nevertheless, the role of pathogenic CNVs in SV and CAVC remain largely unknown because of their low incidence. In our study, genome-wide CNVs in 521 Chinese children with CAVC and SV were screened. A total of 27 CNVs ≥ 200 kb was detected, comprising 10 deletions and 17 duplications, in 11.52% (60/521) CHD cases, namely 16.79% (44/262) in CAVC cases and 6.18% (16/259) in SV cases. According to our strategy, 6 potentially-causative CNVs in 43 cases were identified and contributed to 16.41% (43/262) CAVC patients. Whereas, 6 potentially-causative CNVs in 6 cases were classified which led to the contribution to 2.32% (6/259) SV cases. CNVs in isolate/syndromic CHD patients have been investigated previously, providing a genome-wide (likely) pathogenic CNV burden ranging from 4.3 to 27.9% [52,53,54,55]. In our study, the rate of potentially-causative CNVs in SV cohort is relatively lower, possibly due to the subphenotype difference of CHD and/or the different stringency in variant interpretation standards. Based on previous study, different cardiac subphenotypes showed various enrichment of large CNV events [55], that is, the detection rates in various types of CHD were different. Additionally, some CNVs < 200 kb, which ignored in the present study, may be pathogenic. Furthermore, genome-wide CNVs with a minor allele frequency (MAF) < 1% are usually recognized as an important contributor to CHD [56] and majority non-causative CNVs in our study had a MAF < 1% in DGV database (Tables 2, 3). The conservative CNV analytic methods used in our study, including the restricted focus on CNVs that were absent in DGV, may result in missing some functional CNVs. Further study of these CNVs is still needed to evaluate the clinical implication.
Conclusion
In conclusion, we identified 12 potentially-causative CNVs in 521 CAVC and SV patients, which represented the largest cohort of these two rare CHD types in China. Most CNVs identified in our study were rare (< 1%), occurring just once among the CAVC or SV samples except the 21q11.2–21q22.3 duplication in CAVC cohort. In this study, Chinese CAVC patients were mostly 21 trisomy with DS, which was consistent with the previous reports. Furthermore, it also suggested that there was no race difference in the close correlation between CAVC and DS patients. Combined with the present CNVs reports of CHD and the intolerance of genes within the CNVs regions, our results provided novel genetic evidences that could help clarify the etiology of CHD. Additionally, the potentially-causative CNVs we detected were seldom overlapped with known CHD loci, which implicated that abundant gene involved in heart development and diverse genetic causes of CHD.
Availability of data and materials
Data generated or analyzed during this study are included in this published article and its additional file. The raw data reported in this paper have been deposited in the OMIX, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (https://ngdc.cncb.ac.cn/omix:accession no.OMIX561). CNVs in our study were called based on human assembly GRCh38 (hg38, https://www.ncbi.nlm.nih.gov/assembly/GCA_000001405.27/).
Abbreviations
- CAVC:
-
Complete Atrioventricular Canal
- SV:
-
Single Ventricle
- ToF:
-
Tetralogy of Fallot
- ASD:
-
Atrial septal defect
- CoA:
-
Coarctation of the Aorta
- VSD:
-
Ventricular Septal Defect
- TGA:
-
Transposition of Great Arteries
- PFO:
-
Patent foramen oval
- PDA:
-
Patent ductus arteriosus
References
Hoffman JI, Kaplan SJ. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39(12):1890–900.
van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ, ET AL. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58(21):2241–7.
Khairy P, Ionescu-Ittu R, Mackie AS, Abrahamowicz M, Pilote L, Marelli AJ. Changing mortality in congenital heart disease. J Am Coll Cardiol. 2010;56(14):1149–57.
Cowan JR, Ware SM. Genetics and genetic testing in congenital heart disease. Clin Perinatol. 2015;42(2):373–93.
Gelb BD. Genetic basis of congenital heart disease. Curr Opin Cardiol. 2004;19(2):110–5.
Patel A, Costello JM, Backer CL, Pasquali SK, Hill KD, Wallace AS, et al. Prevalence of noncardiac and genetic abnormalities in neonates undergoing cardiac operations: analysis of the society of thoracic surgeons congenital heart surgery database. Ann Thorac Surg. 2016;102(5):1607–14.
Nikyar B, Sedehi M, Qorbani M, Nikyar A, Golalipour MJ. Ethnical variations in the incidence of congenital heart defects in gorgan, northern iran: a single-center study. 2014, 9(1):9–14.
Warnes CA. Sex differences in congenital heart disease: should a woman be more like a man? Circulation. 2008;118(1):3–5.
Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593–601.
Gifford CA, Ranade SS, Samarakoon R, Salunga HT, de Soysa TY, Huang Y, et al. Oligogenic inheritance of a human heart disease involving a genetic modifier. Science. 2019;364(6443):865-870.
Nees SN, Chung WK. Genetic Basis of Human Congenital Heart Disease. Cold Spring Harb Perspect Biol. 2020;12(9):a036749.
Pierpont ME, Brueckner M, Chung WK, Garg V, Lacro RV, McGuire AL, et al. Genetic basis for congenital heart disease: revisited: a scientific statement from the american heart association. Circulation. 2018;138(21):e653–711.
Mills R, Pittard W, Mullaney J, Farooq U, Creasy T, Mahurkar A, et al. Natural genetic variation caused by small insertions and deletions in the human genome. 2011;21(6):830–9.
Abel H, Duncavage EJ. Detection of structural DNA variation from next generation sequencing data: a review of informatic approaches. 2013;206(12):432–40.
Digilio MC, Romana Lepri F, Dentici ML, Henderson A, Baban A, Roberti MC, et al. Atrioventricular canal defect in patients with RASopathies. Eur J Hum Genet. 2013;21(2):200–4.
Linglart L, Gelb BD. Congenital heart defects in Noonan syndrome: diagnosis, management, and treatment. 2020, 184(1):73–80.
Alongi AM, Kirklin JK, Deng L, Padilla L, Pavnica J, Romp RL, et al. Surgical management of heterotaxy syndrome: current challenges and opportunities. 2020;11(2):166–76.
Liu C, Cao R, Xu Y, Li T, Li F, Chen S, et al. Rare copy number variants analysis identifies novel candidate genes in heterotaxy syndrome patients with congenital heart defects. Genome Med. 2018;10(1):40.
Costain G, Silversides CK, Bassett AS. The importance of copy number variation in congenital heart disease. NPJ Genomic Med. 2016;1(1):16031.
Glessner JT, Bick AG, Ito K, Homsy J, Rodriguez-Murillo L, Fromer M, et al. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ Res. 2014;115(10):884–96.
Dong Z, Zhang J, Hu P, Chen H, Xu J, Tian Q, et al. Low-pass whole-genome sequencing in clinical cytogenetics: a validated approach. 2016;18(9):940–8.
Costain G, Silversides CK, Bassett AS. The importance of copy number variation in congenital heart disease. NPJ Genom Med. 2016;1:16031.
Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, et al. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci USA. 2011;108(7):2915–20.
Sanchez-Castro M, Eldjouzi H, Charpentier E, Busson PF, Hauet Q, Lindenbaum P, et al. Search for rare copy-number variants in congenital heart defects identifies novel candidate genes and a potential role for FOXC1 in Patients With Coarctation of the Aorta. Circ Cardiovasc Genet. 2016;9(1):86–94.
An Y, Duan W, Huang G, Chen X, Li L, Nie C, et al. Genome-wide copy number variant analysis for congenital ventricular septal defects in Chinese Han population. BMC Med Genom. 2016;9:2.
Costain G, Lionel A, Ogura L, Marshall C, Scherer S, Silversides C, et al. Genome-wide rare copy number variations contribute to genetic risk for transposition of the great arteries. 2016;204:115–21.
Silversides CK, Lionel AC, Costain G, Merico D, Migita O, Liu B, et al. Rare copy number variations in adults with tetralogy of Fallot implicate novel risk gene pathways. PLoS Genet. 2012;8(8):e1002843.
Reller MD, Strickland MJ, Riehle-Colarusso T, Mahle WT, Correa A. Prevalence of congenital heart defects in metropolitan Atlanta, 1998–2005. J Pediatr. 2008;153(6):807–13.
Calabrò R, Limongelli G. Complete atrioventricular canal. Orphanet J Rare Dis. 2006;1:8.
Garcia AM, Nakano SJ, Karimpour-Fard A, Nunley K, Blain-Nelson P, Stafford NM, et al. Phosphodiesterase-5 is elevated in failing single ventricle myocardium and affects cardiomyocyte remodeling in vitro. Circ Heart Fail. 2018;11(9):e004571.
Pelleri MC, Cicchini E, Locatelli C, Vitale L, Caracausi M, Piovesan A, et al. Systematic reanalysis of partial trisomy 21 cases with or without Down syndrome suggests a small region on 21q22. 13 as critical to the phenotype. Human Mol Genet. 2016;25(12):2525–38.
Dobosz A, Bik-Multanowski M. Long-term trends in the prevalence of congenital heart defects in patients with Down syndrome in southern Poland. Devel Period Med. 2019;23(3):184–9.
Diogenes TCP, Mourato FA, de Lima Filho JL, da Silva MS. Gender differences in the prevalence of congenital heart disease in Down’s syndrome: a brief meta-analysis. BMC Med Genet. 2017;18(1):111.
Bush D, Galambos C, Ivy D, Abman S, Wolter-Warmerdam K, Hickey F. Clinical characteristics and risk factors for developing pulmonary hypertension in children with down syndrome. 2018;202:212–9.
Pugnaloni F, Digilio MC, Putotto C, De Luca E, Marino B, Versacci P. Genetics of atrioventricular canal defects. Ital J Pediatr. 2020;46:1–13.
Li L, Huang L, Luo Y, Huang X, Lin S, Fang Q. Differing microdeletion sizes and breakpoints in chromosome 7q11.23 in williams-beuren syndrome detected by chromosomal microarray analysis. Mol Syndromol. 2016;6(6):268–75.
Pober B. Williams-Beuren syndrome. 2010;362(3):239–52.
Pober BR, Johnson M, Urban Z. Mechanisms and treatment of cardiovascular disease in Williams-Beuren syndrome. J Clin Investig. 2008;118(5):1606–15.
Verheij J, de Munnik S, Dijkhuizen T, de Leeuw N, Olde Weghuis D, van den Hoek G, et al. An 8.35 Mb overlapping interstitial deletion of 8q24 in two patients with coloboma, congenital heart defect, limb abnormalities, psychomotor retardation and convulsions. Eur J Med Genet. 2009;52(5):353–7.
Dauber A, Golzio C, Guenot C, Jodelka FM, Kibaek M, Kjaergaard S, et al. SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24. 3 copy-number variant. Am J Hum Genet. 2013;93(5):798–811.
Bluhm A, Viceconte N, Li F, Rane G, Ritz S, Wang S, et al. ZBTB10 binds the telomeric variant repeat TTGGGG and interacts with TRF2. Nucleic Acids Res. 2019;47(4):1896–907.
Lahm H, Jia M, Dressen M, Wirth F, Puluca N, Gilsbach R, et al. Genome-wide association study in European patients with congenital heart disease identifies risk loci for transposition of the great arteries and anomalies of the thoracic arteries and veins and expression of discovered candidate genes in the developing heart. bioRxiv 2020.
Nectoux J, Florian C, Delepine C, Bahi-Buisson N, Khelfaoui M, Reibel S, et al. Altered microtubule dynamics in Mecp2-deficient astrocytes. J Neurosci Res. 2012;90(5):990–8.
Radhakrishna U, Albayrak S, Zafra R, Baraa A, Vishweswaraiah S, Veerappa AM, et al. Placental epigenetics for evaluation of fetal congenital heart defects: Ventricular Septal Defect (VSD). PloS one. 2019;14(3):e0200229.
Tomita-Mitchell A, Mahnke DK, Struble CA, Tuffnell ME, Stamm KD, Hidestrand M, et al. Human gene copy number spectra analysis in congenital heart malformations. Physiol Genomics. 2012;44(9):518–41.
Deng X, Zhou J, Li FF, Yan P, Zhao EY, Hao L, et al. Characterization of nodal/TGF-lefty signaling pathway gene variants for possible roles in congenital heart diseases. PLoS One. 2014;9(8):e104535.
Rydeen AB, Waxman JS. Cyp26 enzymes facilitate second heart field progenitor addition and maintenance of ventricular integrity. PLoS Biol. 2016;14(11):e2000504.
Bittel DC, Kibiryeva N, O’Brien JE, Lofland GK, Butler MG. Gene expression in pediatric heart disease with emphasis on conotruncal defects. Prog Pediatr Cardiol. 2005;20(2):127–41.
Uosaki H, Fukushima H, Takeuchi A, Matsuoka S, Nakatsuji N, Yamanaka S, et al. Efficient and scalable purification of cardiomyocytes from human embryonic and induced pluripotent stem cells by VCAM1 surface expression. PLoS One. 2011;6(8):e23657.
Mommersteeg MT, Yeh ML, Parnavelas JG, Andrews WD. Disrupted Slit-Robo signalling results in membranous ventricular septum defects and bicuspid aortic valves. Cardiovasc Res. 2015;106(1):55–66.
Fotiou E, Williams S, Martin-Geary A, Robertson DL, Tenin G, Hentges KE, et al. Integration of large-scale genomic data sources with evolutionary history reveals novel genetic loci for congenital heart disease. Circ Genom Precis Med. 2019;12(10):e002694.
Geng J, Picker J, Zheng Z, Zhang X, Wang J, Hisama F, et al. Chromosome microarray testing for patients with congenital heart defects reveals novel disease causing loci and high diagnostic yield. BMC Genomics. 2014;15(1):1127.
Lee MY, Won HS, Han YJ, Ryu HM, Lee DE, Jeong BD. Clinical value of chromosomal microarray analysis in prenatally diagnosed dextro-transposition of the great arteries. J Matern Fetal Neonatal Med. 2020;33(9):1480–5.
Song T, Wan S, Li Y, Xu Y, Dang Y, Zheng Y, et al. Detection of copy number variants using chromosomal microarray analysis for the prenatal diagnosis of congenital heart defects with normal karyotype. J Clin Lab Anal. 2019;33(1):e22630.
Wu X, Li R, Fu F, Pan M, Han J, Yang X, et al. Chromosome microarray analysis in the investigation of children with congenital heart disease. BMC Pediatr. 2017;17(1):117.
Liu Y, Chang X, Glessner J, Qu H, Tian L, Li D, et al. Association of rare recurrent copy number variants with congenital heart defects based on next-generation sequencing data from family trios. Front Genet. 2019;10:819.
Acknowledgements
We thank the patients and their family members for participating in this study.
Funding
The work was supported by National Natural Science Foundation of China (81801460, 81871717, 81672090) and Shanghai Sailing Program (Grant numbers 18YF1414800), Collaborative Innovation Program of Shanghai Municipal Health Commission (2020CXJQ01), Shanghai Municipal Science and Technology Major Project (2017SHZDZX01), Science and Technology Commission of Shanghai Municipality (No. 20JC1418500, No. 21Y31900301), Shanghai Key Laboratory of Clinical Molecular Diagnostics for Pediatrics (20dz2260900). The funder has no roles in study design, data collection, analysis, and interpretation, in the writing of the manuscript, and in the decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
Conceptualization: QHF & YGY; Investigation: XYZ, XQZ, BW, GLY, YX; Writing of the original draft: XYZ, XQZ; Samples: XYZ; Preparation: XYZ; Review and editing: QHF, YGY& XQZ. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the Ethical Committee of Shanghai Children’s Medical Center (No. SCMCIRB-K2017009). All written informed consents were obtained from the patients or guardian of participants under 16 years old to use their clinical data and samples.
Consent for publication
Not applicable.
Competing interests
None declared.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Table S1. Phenotypes of CHD cohort.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, X., Wang, B., You, G. et al. Copy number variation analysis in Chinese children with complete atrioventricular canal and single ventricle. BMC Med Genomics 14, 243 (2021). https://doi.org/10.1186/s12920-021-01090-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-021-01090-y