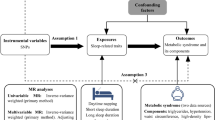
Abstract
Background
Melatonin is a hormone that is secreted at night by the pineal gland. It exerts its function by binding to the MT1 and MT2 receptors, which are encoded by the MTNR1A and MTNR1B genes, respectively. Previous studies reveal that MTNR1B variants are associated with insulin secretion impairments and an increased body mass index (BMI) in individuals of European and Asian ancestries. Obesity is highly prevalent in the US and disproportionately affects African Americans. Here, we hypothesized that common single nucleotide polymorphisms (SNPs) imputed in 1000 Genomes in the MTNR1B gene are associated with adiposity in African American adult men and women and that the association is modified by insomnia.
Methods
We used an additive genetic model to describe the association between the adiposity traits (BMI and waist circumference) and selected MTNR1B variants in 3,029 Jackson Heart Study participants, with an average age of 55.13 ± 12.84 years, and 62% were women. We regressed the adiposity measures on the estimated allelic or genotypic dosage at every selected SNP and adjusted for age, sex, population stratification, and insomnia. Thirty common SNPs, spanning the MTNR1B gene, with a minor allele frequency ≥ 5%, a call rate ≥ 90%, a Hardy–Weinberg equilibrium p value > 10–6, were available for the analysis.
Results
The allele T of rs76371840 was associated with adiposity (OR = 1.47 [1.13—1.82]; PFDR-adjusted = 0.0499), and the allele A of rs8192552 showed a significant association with waist circumference (β = 0.023 ± 0.007; PFDR-adjusted = 0.0077) after correcting for multiple testing. When insomnia was included in the adiposity analysis model, the following four variants became significantly associated with adiposity: rs6483208; rs4388843; rs4601728; and rs12804291.
Conclusions
Our data indicate that polymorphisms in the MTNR1B gene are associated with obesity traits in African Americans. To the best of our knowledge, this is the first study to explore the effect of insomnia on the association between the circadian MTNR1B genetic variants and metabolic traits in an African American sample population. We observed that insomnia affected the association between the MTNR1B variants and adiposity.
Similar content being viewed by others
Background
Melatonin is a chronobiotic hormone that is synthesized by the pineal gland at night [1]. Melatonin is not stored within the pineal gland, but due to its lipophilic nature, it diffuses in the bloodstream, where it rapidly reaches target tissues. Melatonin’s function is exerted in a tissue-specific manner by binding to specific G-coupled protein receptors known as melatonin receptor type 1 (MT1) and melatonin receptor type 2 (MT2). These receptors are present in many tissues and organs [1, 2]. In humans, MT1 and MT2 are encoded by the MTNR1A and MTNR1B genes, respectively.
Multiple studies demonstrate that melatonin is involved in regulating sleep, circadian rhythms, reproduction, and metabolic processes [3,4,5]. Polymorphisms in the MTNR1B gene have been linked to impairments in insulin secretion, fasting blood glucose (FBG) levels, and an increased body mass index (BMI) in individuals of European and Asian ancestry [6,7,8,9]. There are various potential mechanisms by which polymorphisms in the MTNR1B gene might increase FBG levels. It has been reported that the common variant rs10830963, which increases MT2 signaling, might also be associated with an increased risk of developing type 2 diabetes (T2D) [9, 10]. Other studies in individuals of European ancestry show that rare variants in the MTNR1B gene result in reduced or absent MT2 signaling, which is also associated with an increased FBG level and an increased risk of developing T2D [11,12,13].
Obesity is defined as a BMI greater than 30, and it is a growing pandemic that affects 36.5% of US adults [14]. Obesity is prevalent across different ethnic groups, with African Americans having the highest age-adjusted rates [15]. Although African Americans have a higher prevalence of obesity [15] and the highest prevalence of short sleep duration and insomnia compared to other ethnicities [16,17,18], there are no documented studies on the influence of MTNR1B on adiposity or the effect of insomnia on this association. This study aimed to investigate whether selected common genetic variations in the MTNR1B gene reported in previous studies were associated with adiposity in a sample of African American individuals and whether those associations were modified by insomnia.
Results
Study population characteristics
The study population characteristics are displayed in Table 1. Of the available study sample, the average age was 55 years old, and 1871 (62%) were women. The participants had a mean BMI of 32 kg/m2, a mean waist circumference of 101 cm, and a total cholesterol of 198 mg/dL. Also, 679 (29%) participants had diabetes, and the study population had a mean fasting glucose of 100 mg/dL, a mean HbA1c percentage of 6, and a fasting insulin of 19 IU/mL. Additionally, 98 (5%) participants reported insomnia.
The ASSOCIATION of MTNR1B SNPs with BMI and waist circumference
As shown in Table 2, multiple linear regression models identified sixteen MTNR1B variants that showed a nominally significant association (FDR p value ≤ 0.05) with continuous obesity traits (BMI and waist circumference). Fourteen MTNR1B SNPs were inversely and significantly associated with BMI. Ten of these remained significant after adjusting for multiple testing. For waist circumference, sixteen MTNR1B SNPs showed a significant inverse association, while rs8192552 showed a significant direct effect.
Association between MTNR1B variants with adiposity adjusting for insomnia
A multiple logistic regression model identified sixteen MTNR1B variants with a significant association with adiposity (Table 3). After adjusting for insomnia, four new variants, not identified in our previous models, showed a nominally significant association (FDR p value ≤ 0.05) with adiposity (Table 3). Based on the functional annotation of the MTNR1B variants, only two of the variations were missense variants, and we identified variants in other regulatory regions. In the regulatory regions of the MTNR1B introns, two of the variants were in the 3 prime-untranslated region (3’-UTR).
Linkage disequilibrium
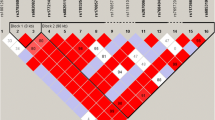
To understand the linkage disequilibrium (LD) around the suggestively or nominally significant MTNR1B variants, we generated an LD plot (Fig. 1). Three haplotypes were identified from the LD plot in the MTNR1B genes from the JHS cohort (Fig. 1). In haplotype block 1, all the variants showed a strong LD (D′ > 1) except for rs76371840. For block 2, all the variants were in strong LD. In haplotype block 3, most variants were in strong LD except rs7130424 & rs37816638 and rs7130424 & rs1562444 (D′ < 1).
Linkage Disequilibrium Plot for SNPs in the MTNR1B gene. The value in each diamond is r2 between pairs of SNPs. The haploview standard color scheme, for LD color display with bright red (LOD ≥ 2 and D′ = 1), shades of pink/red (LOD ≥ 2 and D′ < 1), white (LOD < 2 and D′ < 1), and blue (LOD < 2 and D′ = 1). Haploview generated haplotypes blocks for SNPs in strong LD
Discussion
In this study, we examined the associations of polymorphisms in MTNR1B with obesity traits in a sample of African Americans at JHS. Our principal findings revealed a significant association of rs8192552 with high waist circumference even after adjusting for multiple testing. Furthermore, the T allele of rs76371840 was associated with adiposity even after adjusting for insomnia in the regression model. We also observed four new variants (allele G of rs6483208, allele A of rs4388843, allele G of rs4601728, and allele T of rs12804291) that showed an effect on adiposity outcome after adjusting for insomnia. To the best of our knowledge, we are the first to report this association.
Our results indicated a significant association of rs8192552 with high waist circumference. The variant rs8192552 is a missense variant that has been extensively studied in T2D and shows no association to T2D in European populations [19] and African American populations [20]. However, rs8192552 is significantly associated with BMI and waist circumference in French and Danish populations [19]. Elsewhere, rs8192552 did not show a significant association with BMI, as well as waist circumference [20, 21]. Andersson et al. [19] reported that the missense variant rs61747139 did not display a significant association with BMI and waist circumference. Allele G of rs61747139 has been reported to cause an amino acid change of lysine to arginine. This codon change could affect gene expression, leading to a defect in melatonin signaling pathways. Karamitri et al. [12] observed that rs61747139 decreased β-arrestin-2 recruitment, while rs8192552 did not. The β-arrestins desensitize G-protein-coupled receptors (e.g., MTNR1B) to prevent further stimulation of G proteins and the downstream signaling pathways [22]. The overstimulation of melatonin signaling by variant rs61747139 via β-arrestin-2 could lead to a downstream effect on markers involved in obesity.
The pairwise variants rs12792653 and rs1562444 are associated with both BMI and waist circumference. These SNPs (rs12792653 and rs1562444) are in the 3’-UTR region of the mRNA that follows the stop codon, which contains regulatory regions essential for post-transcription regulation [23]. It also contains binding sites for regulatory elements, such as microRNAs, repressors proteins, and proteins that bind to AU-rich elements (ARE-BP), which are involved in either translation activation or repression [23, 24]. Thus, 3’-UTRs act as cis-regulators.
Additionally, insomnia modified the association between the MTNR1B variants and adiposity because we observed four novel variants (rs6483208, rs4388843, rs4601728, and rs12804291) with a significant association with adiposity when we adjusted for insomnia. To date, few studies have investigated the association between the circadian-related gene variants with metabolic parameters [25,26,27,28]. Ollila et al. described the effect of insomnia on the association of MTNR1B variants on blood glucose [25]. Another study discussed the effect of diet and sleep on the association between circadian-related gene variants (MTNR1B, CLOCK, CRY, and NR1D1) and metabolic traits (fasting glucose, BMI, waist circumference, and HDL-cholesterol) [28]. Our study demonstrated that insomnia affected the association between MTNR1B variants and adiposity in an African American sample population.
While these findings are insightful to the role of MTNR1B variants in African Americans, a few limitations are worth mentioning. Our main limitation is the small sample size relative to studies of different ethnic backgrounds. Our study needs to be replicated in a larger African American cohort. Another limitation is the use of BMI to assess adiposity instead of the overall body fat mass. Despite these limitations, our data produced associations that were similar to other studies.
Conclusions
In summary, we found a novel association between allele A of rs8192552 and high waist circumference. Also, allele T of rs76371840 showed an association with adiposity. Moreover, allele G of rs12792653 and allele G of rs1562444 in the 3’-UTR were associated with BMI and waist circumference. Additionally, when the data was adjusted for insomnia in the adiposity model, allele G of rs6483208, allele A of rs4388843, allele G of rs4601728, and allele T of rs12804291 showed a significant association with adiposity. In conclusion, our study identified several MTNR1B variants associated with obesity in the Jackson Heart Study Population. These findings contribute to understanding the link between circadian disruption (insomnia) and metabolic homeostasis.
Methods
Study subject
For this study, we used cross-sectional data from the Jackson Heart Study (JHS). The JHS is a single-site community-based cohort study of risk factors for cardiovascular disease among adult African American men and women living in the Jackson, Mississippi, Metropolitan area. The study participants consisted of 5,306 individuals recruited, interviewed, and examined by certified technicians for the first exam (2000–2004) [29, 30] and were followed up in 2 subsequent exams from 2005 to 2008 and 2009 to 2013. The clinic visits encompassed a physical examination, blood and urine collection, anthropometry, and data collection regarding family history, behavioral risk factors, and sociodemographics. There were 3027 participants (Mean age of 55.13 ± 12.84 and 1871 women) who consented to the genetic analysis, and their DNA samples were genotyped in the candidate gene association resource (CARe) consortium using the Affymetrix 6.0 platform. They were later imputed to 1000 genomes phase 1 [31,32,33]. The study was approved by the Institutional Review Board (IRB) of the National Institutes of Health. The IRB approved the participating institutions' protocol (University of Mississippi Medical Center, Jackson State University, and Tougaloo College).
Study variables
Outcome variables
Our primary outcome in this study is adiposity (BMI and waist circumference). Adiposity was measured in exam visit 1, and we defined it as a BMI greater than 30 kg/m2 and a waist circumference greater than 102 cm and 88 cm for men and women, respectively. All the above clinical parameters were measured according to standard laboratory and clinical techniques [30].
Independent variables: SNP selection genotyping and imputation
All 3027 JHS samples were genotyped on the Affymetrix 6.0 based on manufacturer protocol [33]. The candidate gene approach was used to select our genetic variants from the entire set of common genetic variants in the MTNR1B gene located on chromosome 11q14.3 and hg19 position base-pair ordinates chr11:92,702,789–92,718,2 (plus-strand orientation). The JHS coordination centers performed the SNPs quality control, and the variants that passed were imputed with 1000G phase 1 using Cosmopolitan reference panel including all races—version 2010–11 data freeze, 2012–03-04 haplotypes [32, 33]. The imputation was completed using Minimac3 on the Michigan Imputation Server [34]; details regarding the reference panel can be found in the 1000 Genomes Project Consortium 2010 [35]. Imputed SNPs were filtered for minor allele frequency ≥ 1%, call rate ≥ 90%, HWE p value > 10–6, as well as the exclusion of sites with invalid or mismatched alleles for the reference panel [32]. For this study, 109 SNPs were genotyped and imputed; we focused on common variants with a minor allele frequency (MAF) ≥ 5%, and with imputation quality ≥ 80%, 30 common variants were selected for downstream analyses. Covariates were age, gender, and 10 principal components to adjust for population stratification due to admixture [17]. In additional analyses, we also adjusted for insomnia covariate. The participants were asked if they have insomnia with the answer option of “Yes,” “No,” and “Don’t Know.” Insomnia is clinically defined as the difficulty of falling and staying asleep [36].
Statistical analysis
Descriptive statistics
The study variables were summarized using the mean and standard deviation (SD) for the continuous variables and proportions for the categorical variables. The continuous variables were first assessed for normality, and then were log-transformed if they were not normally distributed. The analyses for the descriptive statistics were performed using the statistical software SAS® 9.4 [37].
Regression analysis
Multivariate logistic regression models were fitted to assess the associations between the dosage of the MTNR1B genetic variants and adiposity after adjusting for age, gender, and the 10 principal components in the adiposity model. Due to the relationship between melatonin signaling and sleep, we examined the modifying effect of insomnia (adiposity-insomnia model) on our adiposity outcome by stratifying each modifier. Multivariate linear regression models (BMI model and waist circumference model) were fitted to examine the relationships between the MTNR1B genetic variants and the continuous obesity outcome traits of BMI and waist circumference. The linear and logistic regression models were fitted using ProbAbel v.0.5.0 genetic analysis software [38], assuming a population-based design. Although a small subset of the JHS participants belonged to a family component, we did not adjust for family structure because previous studies have shown a minimal impact on power and the inflation of the type I error [39,40,41,42]. We used a false discovery rate (FDR) to correct for multiple testing with an adjusted p value threshold of 0.05. The NIH dbSNP database was used to annotate the function of the MTNR1B variants that displayed a significant association using the regression models [43].
The variants that were statistically significant after the FDR adjustment were used to generate a linkage disequilibrium (LD) plot. Haploview (Broad Institute, MA, USA) was used to create the LD plot, and we used the Yoruba, Nigeria population as a reference [44]. Haploview generated the haplotype blocks in the LD plot whenever 95% of the informative comparisons were in strong LD while ignoring variants with an MAF < 0.05 [45].
Availability of data and materials
All the data in the present study required prior approval of a manuscript proposal by the Jackson Heart Study Presentation and Publications and Sub-Committee and a signed Jackson Heart Study data use agreement; Publication ID: P0946. The Jackson Heart Study provides all the details for the data access request (https://www.jacksonheartstudy.org/Research/Study-Data/Data-Access).
Abbreviations
- BMI:
-
Body mass index
- MTNR1A:
-
Melatonin receptor type 1 gene
- MTNR1B:
-
Melatonin receptor type 2 gene
- FDR:
-
False discovery rate
- HWE:
-
Hardy–Weinberg equilibrium
- JHS:
-
Jackson Heart Study
- LD:
-
Linkage disequilibrium
- LCI:
-
Lower confidence interval
- MAF:
-
Minor allele frequency
- UCI:
-
Upper confidence interval
- SE:
-
Standard error
- SNP:
-
Single nucleotide polymorphisms
- OR:
-
Odd ratio
- T2D:
-
Type 2 diabetes
References
Jockers R, Delagrange P, Dubocovich ML, Markus RP, Renault N, Tosini G, et al. Update on melatonin receptors: IUPHAR Review 20. Br J Pharmacol. 2016;173(18):2702–25.
Brydon L, Roka F, Petit L, de Coppet P, Tissot M, Barrett P, et al. Dual signaling of human mel1a melatonin receptors via Gi2, Gi3, and Gq/11 proteins. Mol Endocrinol. 1999;13(12):2025–38. https://doi.org/10.1210/mend.13.12.0390.
Xie Z, Chen F, Li WA, Geng X, Li C, Meng X, et al. A review of sleep disorders and melatonin. Neurol Res. 2017;39(6):559–65. https://doi.org/10.1080/01616412.2017.1315864.
Favero G, Stacchiotti A, Castrezzati S, Bonomini F, Albanese M, Rezzani R, et al. Melatonin reduces obesity and restores adipokine patterns and metabolism in obese (ob/ob) mice. Nutr Res. 2015;35(10):891–900.
Owino S, Buonfiglio DDC, Tchio C, Tosini G. Melatonin signaling a key regulator of glucose homeostasis and energy metabolism. Front Endocrinol (Lausanne). 2019. https://doi.org/10.3389/fendo.2019.00488.
Been LF, Hatfield JL, Shankar A, Aston CE, Ralhan S, Wander GS, et al. A low frequency variant within the GWAS locus of MTNR1B affects fasting glucose concentrations: genetic risk is modulated by obesity. Nutr Metab Cardiovasc Dis. 2012;22(11):944–51.
Liu CT, Raghavan S, Maruthur N, Kabagambe EK, Hong J, Ng MCY, et al. Trans-ethnic meta-analysis and functional annotation illuminates the genetic architecture of fasting glucose and insulin. Am J Hum Genet. 2016;99(1):56–75.
Karamitri A, Jockers R. Melatonin in type 2 diabetes mellitus and obesity. Nat Rev Endocrinol. 2019;15:105–25.
Tuomi T, Nagorny CLF, Singh P, Bennet H, Yu Q, Alenkvist I, et al. Increased melatonin signaling is a risk factor for type 2 diabetes. Cell Metab. 2016;23(6):1067–77.
Lyssenko V, Nagorny CLF, Erdos MR, Wierup N, Jonsson A, Spégel P, et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41(1):82–8.
Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, Sparso T, Holmkvist J, Marchand M, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41(1):89–94. https://doi.org/10.1038/ng.277.
Karamitri A, Plouffe B, Bonnefond A, Chen M, Gallion J, Guillaume J-L, et al. Type 2 diabetes–associated variants of the MT2 melatonin receptor affect distinct modes of signaling. Sci Signal. 2018;11(545):eaan6622.
Bonnefond A, Clément N, Fawcett K, Yengo L, Vaillant E, Guillaume J-L, et al. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet. 2012;44(3):297–301.
Overweight & Obesity | CDC [Internet]. [cited 2020 May 28]. Available from: https://www.cdc.gov/obesity/index.html
Petersen R, Pan L, Blanck HM. Racial and ethnic disparities in adult obesity in the United States: CDC’s tracking to inform state and local action. Prev Chronic Dis. 2019. https://doi.org/10.5888/pcd16.180579.
Williams NJ, Grandner MA, Snipes SA, Rogers A, Williams O, Airhihenbuwa C, et al. Racial/ethnic disparities in sleep health and health care: importance of the sociocultural context. vol 1, Sleep health. Elsevier Inc.; 2015. p. 28–35.
Slopen N, Lewis TT, Williams DR. Discrimination and sleep: A systematic review. Vol. 18, Sleep Medicine. Elsevier; 2016. p. 88–95.
Ruiter ME, DeCoster J, Jacobs L, Lichstein KL. Sleep disorders in African Americans and Caucasian Americans: a meta-analysis. Behav Sleep Med. 2010;8(4):246–59.
Andersson EA, Holst B, Sparsø T, Grarup N, Banasik K, Holmkvist J, et al. MTNR1B G24E variant associates with BMI and fasting plasma glucose in the general population in studies of 22,142 Europeans. Diabetes. 2010;59(6):1539–48.
Ng MCY, Hester JM, Wing MR, Li J, Xu J, Hicks PJ, et al. Genome-wide association of BMI in African Americans. Obesity. 2012;20(3):622–7. https://doi.org/10.1038/oby.2011.154.
Monda KL, Chen GK, Taylor KC, Palmer C, Edwards TL, Lange LA, et al. A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet. 2013;45(6):690–6.
Gainetdinov RR, Premont RT, Bohn LM, Lefkowitz RJ, Caron MG. Desensitization of G protein-coupled receptors and neuronal functions. Annu Rev Neurosci. 2004;27(1):107–44. https://doi.org/10.1146/annurev.neuro.27.070203.144206.
Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci. 2012;69(21):3613–34.
Pichon X, Wilson LA, Stoneley M, Bastide A, King HA, Somers J, et al. RNA binding protein/RNA element interactions and the control of translation. Curr Protein Pept Sci. 2012;13(4):294–304.
Ollila HM, Kronholm E, Kettunen J, Silander K, Perola M, Porkka-Heiskanen T, et al. Insomnia does not mediate or modify the association between MTNR1B risk variant rs10830963 and glucose levels. Vol. 59, Diabetologia. Springer Verlag; 2016. p. 1070–2.
Silva ACP, Santos MJ, Koike BDV, Moreira MSA, Gitai DLG, de Miranda Coelho JAP, et al. Melatonin receptor 1B–1193T>C polymorphism is associated with diurnal preference and sleep habits. Sleep Med. 2019;53:106–14.
Lane JM, Chang AM, Bjonnes AC, Aeschbach D, Anderson C, Cade BE, et al. Impact of common diabetes risk variant in MTNR1B on sleep, circadian, and melatonin physiology. Diabetes. 2016;65(6):1741–51.
Dashti HS, Follis JL, Smith CE, Tanaka T, Garaulet M, Gottlieb DJ, et al. Gene-environment interactions of circadian-related genes for cardiometabolic traits. Diabetes Care. 2015;38(8):1456–66.
Taylor HA. The Jackson Heart Study: an overview. Ethn Dis. 2005;15(4 Suppl 6):S6-1–3.
Carpenter MA, Crow R, Steffes M, Rock W, Skelton T, Heilbraun J, et al. Laboratory, reading center, and coordinating center data management methods in the Jackson Heart Study. Am J Med Sci. 2018;328(3):131–44. https://doi.org/10.1097/00000441-200409000-00001.
Musunuru K, Lettre G, Young T, Farlow DN, Pirruccello JP, Ejebe KG, et al. Candidate Gene Association Resource (CARe) design, methods, and proof of concept. Circ Cardiovasc Genet. 2010;3(3):267–75.
Duan Q, Liu EY, Auer PL, Zhang G, Lange EM, Jun G, et al. Imputation of coding variants in African Americans: better performance using data from the exome sequencing project. Bioinformatics. 2013;29(21):2744–9. https://doi.org/10.1093/bioinformatics/btt477.
Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ, et al. Genome-Wide Association Study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe project. PLoS Genet. 2011;7(2):e1001300.
Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–7.
Auton A, Abecasis GR, Altshuler DM, Durbin RM, Bentley DR, Chakravarti A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
Bonnet MH, Arand DL. Chronic insomnia. In: Sleep Disorders: Diagnosis and Therapeutics. CRC Press; 2008. p. 80–9.
SAS ® 9.4 Statements Reference SAS ® Documentation. 2013.
Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinform. 2010;11(1):134. https://doi.org/10.1186/1471-2105-11-134.
Liu S, Wilson JG, Jiang F, Griswold M, Correa A, Mei H. Multi-variant study of obesity risk genes in African Americans: the Jackson Heart Study. Gene. 2016;593(2):315–21.
McArdle PF, O’Connell JR, Pollin TI, Baumgarten M, Shuldiner AR, Peyser PA, et al. Accounting for relatedness in family based genetic association studies. Hum Hered. 2007;64(4):234–42.
Wang K, Hu X, Peng Y. An analytical comparison of the principal component method and the mixed effects model for association studies in the presence of cryptic relatedness and population stratification. Hum Hered. 2013;76(1):1–9.
Musani SK, Fox ER, Kraja A, Bidulescu A, Lieb W, Lin H, et al. Genome-wide association analysis of plasma B-type natriuretic peptide in blacks: the Jackson Heart Study. Circ Cardiovasc Genet. 2015;8(1):122–30.
Kitts A, Sherry S. The Single Nucleotide Polymorphism Database (dbSNP) of Nucleotide Sequence Variation. 2011;
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. https://doi.org/10.1093/bioinformatics/bth457.
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–9.
Acknowledgements
The authors wish to thank the staff and participants of the Jackson Heart Study for their long-term commitment and important contributions to the study.
Funding
The Jackson Heart Study (JHS) is supported and conducted in collaboration with Jackson State University (HHSN268201800013I), Tougaloo College (HHSN268201800014I), the Mississippi State Department of Health (HHSN268201800015I), and the University of Mississippi Medical Center (HHSN268201800010I, HHSN268201800011I, and HHSN268201800012I) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute on Minority Health and Health Disparities (NIMHD). This study was supported by the National Institutes of Health (NIH) Grant T-32 HL103104 and R01EY026291to G.T; and by 5U54NS083932, S21MD000101, G12-RR03034, U54RR026137 to Morehouse School of Medicine.
Author information
Authors and Affiliations
Contributions
CT and GT conceived the study. CT and SM performed the analyses.AQ assisted in the data analyses and manuscript revision. CT, GT, AQ, and SM wrote the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethics approval and written consent were obtained from each participant before the data collection. The study protocol was approved by the Institutional Review Boards of the National Institutes of Health and the participating Jackson Heart Study institutions, including the University of Mississippi Medical Center, Tougaloo College, and Jackson State University. The final version of the manuscript was approved by the Jackson Heart Study Publications and Presentations Subcommittee.
Consent for publication
Not applicable.
Competing interests
The author(s) declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tchio, C., Musani, S.K., Quarshie, A. et al. Association between MTNR1B polymorphisms and obesity in African American: findings from the Jackson Heart Study. BMC Med Genomics 14, 136 (2021). https://doi.org/10.1186/s12920-021-00983-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-021-00983-2