Abstract
Background
Mexico is among the countries showing the highest heterogeneity of CFTR variants. However, no de novo variants have previously been reported in Mexican patients with cystic fibrosis (CF).
Case presentation
Here, we report the first case of a novel/de novo variant in a Mexican patient with CF. Our patient was an 8-year-old male who had exhibited the clinical onset of CF at one month of age, with steatorrhea, malabsorption, poor weight gain, anemia, and recurrent respiratory tract infections. Complete sequencing of the CFTR gene by next generation sequencing (NGS) revealed two different variants in trans, including the previously reported CF-causing variant c.3266G > A (p.Trp1089*, W1089*), that was inherited from the mother, and the novel/de novo CFTR variant c.1762G > T (p.Glu588*).
Conclusion
Our results demonstrate the efficiency of targeted NGS for making a rapid and precise diagnosis in patients with clinically suspected CF. This method can enable the provision of accurate genetic counselling, and improve our understanding of the molecular basis of genetic diseases.
Similar content being viewed by others
Background
Cystic fibrosis (CF, MIM# 219700) is the most common autosomal recessive disorder among Caucasians. Its incidence varies between different populations, spannig from 1/900 to 1/25,000 or even lower in Eastern populations [1].
Despite the recent instigation of neonatal screening, molecular diagnosis of CF is challenging due to the complex genotype–phenotype relationship, and high genetic heterogeneity [2,3,4], as found in the Latino American population [5,6,7]. Since the CFTR gene was identified as being responsible for CF [8], over 2000 variants have been detected, with the deletion of phenylalanine at position 508 (c.1521_1523delCTT, p.Phe508del, F508del) being the most frequent worldwide [9]. To date, only nine reported CF cases have involved de novo variants [10,11,12,13,14,15]. Although Mexico is among the countries showing the highest heterogeneity of CFTR pathogenic variants, no de novo variants have previously been reported in Mexican patients.
Here, we report the first case of a novel/de novo variant in a Mexican patient with CF.
Case presentation
Clinical phenotype
An 8-year-old male Mexican patient was referred to our Institution with a diagnosis of CF. He was the fifth child born from healthy non-consanguineous parents, without a family history of the disease (Fig. 1). The mother previously had a spontaneous abortion after 16 weeks gestation by anencephaly. Clinical onset occurred at one month of age with steatorrhea, malabsorption, poor weight gain, and anemia. At 5 months of age, the patient had recurrent respiratory tract infections colonized by Pseudomonas aeruginosa. He was diagnosed with CF at 18 months of age, with elevated sweat chloride levels (88, 130, and 129 mmol/l). Currently, the patient’s weight is below the 5th percentile and his height is between the 5th and 10th percentiles.
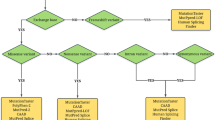
a Family pedigree showing the carriers of variants c.3266G > A, p.Trp1089* and c.1762G > T, p.Glu588*, and the patient with both variants (p.Trp1089*/p.Glu588*). b Sanger sequencing of the patient, father, and mother. Left, sequence of the variant c.3266G > A, p.Trp1089*. Right, sequence of the variant c.1762G > T, p.Glu588*
Molecular analysis
Using samples from the patient, his parents, and his four siblings, we extracted genomic DNA from peripheral blood lymphocytes using the QIAamp DNA Blood Maxi kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol. The index case was screened for the five pathogenic variants that are most frequent in the Mexican population (c.1521_1523delCTT, p.Phe508del; c.1624G > T, p.Gly542*; c.1519_1521delATC, p.Ile507del; c.1646G > A, p.Ser549Asn; and c.3909C > G, p.Asn1303Lys [16,17,18]), using PCR-mediated site-directed mutagenesis (PSM) as previously reported [19, 20]. Since none of the above-listed variants were identified, we performed complete sequencing of the CFTR gene using next generation sequencing (NGS; Illumina HiSeq 2500 sequencer). The NGS data were analyzed using the Genome Analysis Toolkit: UnifiedGenotyper (GATK) and Integrative Genomics Viewer (IGV) software. To predict the functional effect of the variant on the protein, we performed in silico analysis using Variant Effect Predictor (VEP) software [21].
Data collection and molecular analysis were approved by the Research and Ethics Committees at INMEGEN. The parents provided written consent for the child participants, and all children assented.
Our initial screening of the patient revealed none of the pathogenic variants that are most frequent in the Mexican population. Analysis of the completely sequenced CFTR gene using GATK and IGV software revealed that the patient carried two different variants in trans. One was c.3266G > A (p.Trp1089*, W1089*), which is a previously report CF-causing variation, and the other was the novel variant c.1762G > T, p.Glu588* (https://www.cftr2.org/, [22]). The novel variation involved the change of a G for T in codon 588 of exon 12. Functional effect prediction analysis revealed that this variant induced a premature stop codon (c.1762G > T, p.Glu588*).
Both variants were validated via automated Sanger sequencing. To identify their carrier status, we screened both parents for these variants. The patient’s mother carried only the c.3266G > A (p.Trp1089*, W1089*) variant, while the father carried neither of the variants identified in the patient (Fig. 1). Since the father was not a carrier of any CF variant, paternity was tested using 23 STR genetic markers, which confirmed a paternal probability of > 99.99%. Thus, c.1762G > T, p.Glu588* was determined to be a de novo variant. The patient’s three sisters were carriers of the c.3266G > A, p.Trp1089* variant, while his brother showed a wild-type CFTR sequence. Additionally, two maternal uncles and the grandmother were also found to carry the c.3266G > A, p.Trp1089 variant.
Discussion and conclusions
To date, only nine cases of CF caused by de novo pathogenic variants have been reported [10,11,12,13,14]. Most of these variants are single-nucleotide variants, although small and large deletions and partial duplications have also been described [14, 15]. The small number of de novo variants reported in recessive disorders is likely due to the extremely low probability of a de novo variant occurring in combination with the transmission of an allele carrying a pathogenic variation from the other parent. It is also possible that traditional technologies did not enable prior detection of these variants. High-throughput NGS technologies have greatly improved the possibility of identifying rare CFTR variants, and of thus further elucidating the genetic heterogeneity of CF [9, 17, 23]. These approaches also dramatically increase the possibility of identifying novel and de novo variants.
In the presently reported case, we identified a novel/de novo variant at position 588, which resulted in a premature stop codon (c.1762G > T, p.Glu588*) in exon 12 of the CFTR gene. The patient was an 8-year-old Mexican male who had experienced the clinical onset of CF at one month of age, with pancreatic insufficiency and obstructive lung disease. Compared to inherited variants, de novo variants are probably more deleterious because they have been subjected to less stringent evolutionary selection [24]. However, our patient also harbored the previously reported variant c.3266G > A (p.Trp1089*, W1089*), which is a severe variant that also introduces a stop codon, UAG, at position 1089 in exon 20 [25]. In silico prediction indicated that both identified variants could lead to the production of a truncated protein, decreased translation and elongation accuracy, or production of a final protein that lacks an exon due to exon skipping [26]. Further functional characterization of the novel variant is necessary [3, 27].
The patient carried two class I variants, and thus would be expected to show severe disease, which is in accordance with the observed phenotype. Family analysis revealed that the patient’s mother, three sisters, two uncles, and grandmother were carriers of the c.3266G > A, p.Trp1089* pathogenic variant. However, the c.1762G > T, p.Glu588* variant was not found in the father or in the siblings, and was thus determined to be a novel and de novo variant in our patient with CF. Since our patient harbors two nonsense pathogenic variants, personalized therapy based on the readthrough approach may be appropriate as a future approach [28].
Interestingly, the previously reported novel variants have been located on the paternal chromosome, as in our present case, supporting reports that the male germ line may be more mutagenic [29, 30]. This assumption is related to paternal age at conception, which correlates with the pathogenic variant rate in germline cells. There is presently insufficient information to assess this possibility in CF, since of the nine previously reported CF cases with de novo variants, only two provide information regarding the father’s age (25 and 32 years old). The father of our present patient was 40 years of age.
The present case report demonstrates the efficiency of using targeted NGS to make a rapid and precise diagnosis in patients with clinically suspected CF. This method facilitates the identification of rare or exclusive variants, supporting the provision of accurate genetic counselling and personalized therapy, as well as furthering our understanding of the molecular basis of genetic diseases.
Abbreviations
- CF:
-
Cystic fibrosis
- CFTR :
-
Cystic fibrosis transmembrane conductance regulator
- GATK:
-
Genome Analysis Toolkit
- IGV:
-
Integrative Genomics Viewer
- INMEGEN:
-
Instituto Nacional de Medicina Genómica
- NGS:
-
Next generation sequencing
- VEP:
-
Variant Effect Predictor
References
Silva Filho LVRF, Castaños C, Ruíz HH. Cystic fibrosis in Latin America improving the awareness. J Cyst Fibros. 2016;15:791–3.
Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16:45–56.
Terlizzi V, Castaldo G, Salvatore D, Lucarelli M, Raia V, Angioni A, et al. Genotype-phenotype correlation and functional studies in patients with cystic fibrosis bearing CFTR complex alleles. J Med Genet. 2017;54:224–35.
Lucarelli M, Bruno SM, Pierandrei S, Ferraguti G, Testino G, Truglio G. The impact on genetic testing of mutational patterns of CFTR gene in different clinical macrocategories of cystic fibrosis. J Mol Diagn. 2016;18:554–65.
Chávez-Saldaña M, Yokoyama E, Lezana JL, Carnevale A, Macías M, Vigueras RM, et al. CFTR allelic heterogeneity in Mexican patients with cystic fibrosis: implications for molecular screening. Rev Investig Clin. 2010;62:546–52.
Nunes LM, Ribeiro R, Niewiadonski VDT, Sabino E, Yamamoto GL, Bertola DR, et al. A new insight into CFTR allele frequency in Brazil through next generation sequencing. Pediatr Pulmonol. 2017;52:1300–5.
Pepermans X, Mellado S, Chialina S, Wagener M, Gallardo L, Lande H, et al. Identification and frequencies of cystic fibrosis mutations in Central Argentina. Clin Biochem. 2016;49:154–60.
Riordan JR, Rommens JM, Kerem B, NO a A, Rozmahel R, Grzelczak Z, et al. Identification the cystic fibrosis Gene : cloning and characterization of complementary DNA. Science. 1989;245:1066–73.
Ivanov M, Matsvay A, Glazova O, Krasovskiy S, Usacheva M, Amelina E, et al. Targeted sequencing reveals complex, phenotype-correlated genotypes in cystic fibrosis. BMC Med Genet. 2018;11:13.
White MB, Leppert M, Nielsen D, Zielenski J, Gerrard B, Stewart C, et al. A de novo cystic fibrosis mutation: CGA (Arg) to TGA (stop) at codon 851 of the CFTR gene. Genomics. 1991;11:778–9.
Cremonesi L, Cainarca S, Rossi A, Padoan R, Ferrari M. Detection of a de novo R1066H mutation in an Italian patient affected by cystic fibrosis. Hum Genet. 1996;98:119–21.
Casals T, Ramos MD, Giménez J, Nadal M, Nunes V, Estivill X. Paternal origin of a de novo novel CFTR mutation (L1065R) causing cystic fibrosis. Hum Mutat. 1988;(Suppl 1):S99-102.
Křenková P, Piskáčková T, Holubová A, Balaščaková M, Krulišová V, Čamajová J, et al. Distribution of CFTR mutations in the Czech population: positive impact of integrated clinical and laboratory expertise, detection of novel/de novo alleles and relevance for related/derived populations. J Cyst Fibros. 2013;12:532–7.
Girodon E, Des georges M, Medina R, Grenet D, Férec C, Claustres M, et al. Ocurrence of CFTR de novo mutations is not so rare. J Cyst Fibros. 2008;7:S6.
Norek A, Stremska M, Sobczyńska-Tomaszewska A, Wertheim-Tysarowska K, Dmeńska H, Jurek M. Novel de novo large deletion in cystic fibrosis transmembrane conductance regulator gene results in a severe cystic fibrosis phenotype. J Pediatr. 2011;159:343–6.
Orozco L, Velázquez R, Zielenski J, Tsui LC, Chávez M, Lezana JL, Saldaña Y, Hernández E, Carnevale A. Spectrum of CFTR mutations in Mexican cystic fibrosis patients: identification of five novel mutations (W1089C, 846delT, P750L, 4160insGGGG and 297-1G--A). Hum Genet. 2000;106:360–5.
Bergougnoux A, D'Argenio V, Sollfrank S, Vermeau F, Telese A, et al. Multicenter validation study for the certification of a CFTR gene scanning method using next generation sequencing technology. Clin Chem Lab Med. 2018;56:1046–53.
Dequeker E, et al. Best practice guidelines for molecular genetic diagnosis of cystic fibrosis and CFTR-related disorders – updated European recommendations. Eur J Hum Genet. 2009;17:51–65.
Friedman KJ, Edward Highsmith W, Silverman LM. Detecting multiple cystic fibrosis mutations by polymerase chain reaction-mediated site-directed mutagenesis. Clin Chem. 1991;37:753–5.
Orozco L, Friedman K, Chávez M, Lezana JL, Villareal MT, Carnevale A. Identification of the I507 deletion by site-directed mutagenesis. Am J Med Genet. 1994;15:137–9.
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The Ensembl variant effect predictor. Genome Biol. 2016;17:1–14.
Claustres M, Thèze C, des Georges M, Baux D, Girodon E, Bienvenu T, et al. CFTR-France, a national relational patient database for sharing genetic and phenotypic data associated with rare CFTR variants. Hum Mutat. 2017;38:1297–315.
Lucarelli M, Porcaro L, Biffignandi A, Costantino I, Gionnone V, et al. A new targeted CFTR mutation panel based on next-generation sequencing technology. J Mol Diagn. 2017;19:788–800.
Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–75.
Shoshanl T, Augarten A, Yahav J, Gazit E, Kerem B. Two novel mutations in the CFTR gene: W1089X in exon 17B and 4010delTATT in exon 21. Hum Mol Genet. 1994;3:657–8.
Kellermayer R. Translational readthrough induction of pathogenic nonsense mutations. Eur J Med Genet. 2006;49:445–50.
Hinzpeter A, Reboul MP, Callebaut I, Zordan C, Costes B, Guichoux J, et al. The importance of functional test to assess the effect of a new CFTR variant when genotype-phenotype correlation is not possible. Clin Case Rep. 2017;5:658–63.
Altamura N, Castaldo R, Finotti A, Breveglieri G, Salvatori F, et al. Tobramycin is a suppressor of premature termination codons. J Cyst Fibrosis. 2013;12:806–11.
Girard SL, Bourassa CV, Lemieux Perreault L-P, Legault M-A, Barhdadi A, Ambalavanan A, et al. Paternal age explains a major portion of De novo germline mutation rate variability in healthy individuals. PLoS One. 2016;11:e0164212.
Rahbari R, Wuster A, Lindsay SJ, Hardwick RJ, Alexandrov LB, Al Turki S, et al. Timing, rates and spectra of human germline mutation. Nat Genet. 2016;48:126–33.
Acknowledgements
We thank the patient and his family for participating in the study. Also, we would like to acknowledgment to M. en C. Alfredo Mendoza and Quim. Juan Luis Jiménez for technical support.
Funding
Julieta Larrosa received a scholarship (#819707) from Consejo Nacional de Ciencia y Tecnología (CONACYT). She is a student of Master’s degree in science of the Maestría y Doctorado en Ciencias Biológicas, Universidad Nacional Autónoma de México (UNAM). The student participated in the analysis and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available.
from the corresponding author on request.
Author information
Authors and Affiliations
Contributions
LO, AMH, conceived and conducted the study; EMC, JLL identified.the patients and carried out the clinical characterizations; JL,FBO, HGO, EM carried. Out the molecular genetics studies; JL, CCC, HGO analysis; LO, AMH, JL wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Data collection and molecular analysis were approved by the Research and Ethics Committees at Instituto Nacional de Medicina Genómica (INMEGEN).
Consent for publication
The parents signed a letter of consent to participate in the study and all children assented. They also gave their authorization for the publication of the results.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Martínez-Hernández, A., Larrosa, J., Barajas-Olmos, F. et al. Next-generation sequencing for identifying a novel/de novo pathogenic variant in a Mexican patient with cystic fibrosis: a case report. BMC Med Genomics 12, 68 (2019). https://doi.org/10.1186/s12920-019-0528-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-019-0528-1