Abstract
Adipogenesis is the process of cell differentiation by which mesenchymal stem cells become adipocytes. Extensive research is ongoing to identify genes, their protein products, and microRNAs that correlate with fat cell development. The existing databases have focused on certain types of regulatory factors and interactions. However, there is no relationship between the results of the experimental studies on adipogenesis and these databases because of the lack of an information center. This information fragmentation hampers the identification of key regulatory genes and pathways. Thus, it is necessary to provide an information center that is quickly and easily accessible to researchers in this field. We selected and integrated data from eight external databases based on the results of text-mining, and constructed a publicly available database and web interface (URL: http://210.27.80.93/arn/), which contained 30873 records related to adipogenic differentiation. Then, we designed an online analysis tool to analyze the experimental data or form a scientific hypothesis about adipogenesis through Swanson’s literature-based discovery process. Furthermore, we calculated the “Impact Factor” (“IF”) value that reflects the importance of each node by counting the numbers of relation records, expression records, and prediction records for each node. This platform can support ongoing adipogenesis research and contribute to the discovery of key regulatory genes and pathways.
Similar content being viewed by others
Background
Adipose tissue is an important site for lipid storage, energy homeostasis, and whole-body insulin sensitivity. It is important to understand the mechanisms involved in adipose tissue development. Growth of adipose tissue is the result of differentiation of new fat cells from precursor cells [1]. It is obvious that adipogenesis is not a single gene trait, but is determined by a number of genes and their encoded proteins [2]. Therefore, researchers need a professional comprehensive knowledge database including related genes, proteins, properties, biological processes, and environmental factors in accordance with their determined or predicted relations in the literature to assist researchers in understanding adipogenic differentiation from the perspective of systems biology.
After obtaining a large amount of data and information related to fat, a key element is linking the extracted information together to form new facts or hypotheses to be explored further by more conventional means of experimentation [3]. Swanson developed and implemented a novel tool to mine the existing knowledge base for unreported or underreported relationships, and highlighted previously published but neglected hypotheses, a process known as literature-based discovery [4]. This process functions by connecting two seemingly unrelated findings [5]. This and implemented a novel tool to mine the existing knowledge and easily accessible to researchers in this field. Conclusive proof, the discovery is, in itself, very helpful to uncover previously unknown relationships [6]. Furthermore, it can help investigators access context and mine knowledge that might not be revealed using a traditional search.
In the present study, we constructed a molecular regulatory network of adipogenesis based on text-mining and manual examination, and then screened the data of four external databases according to the network, which produced more than 10 000 prediction results out of >1 × 106 interaction records (Table 1). Moreover, we designed an online analysis tool according to the theory of “literature-based discovery”, which provides a new approach for researchers to analyze data and form hypotheses. Ultimately, we explored the possibility of using a large amount of accumulated data to promote future research and practices.
Construction and content
The Adipogenic Regulation Network (ARN) Database aims to provide a high-quality collection of genes, microRNAs, and their relations implicated in the regulation of adipogenesis, which has been reviewed by experts in the field. The data collection and processing steps are illustrated in Fig. 1. The workflow included four major steps. Step one: build a text-mining association network using the Agilent Literature Search plugin [7]. Step two: information processing and analysis. Step three: screen the data of four external databases (Table 1) according to the network. Step four: design analysis tool.
Information mining and manual review
For the literature search, we established a set of queries by entering 47 key genes in adipogenesis [7] with simultaneous input contexts ‘adipo* differen*’, which is short for “adipocyte differentiation”. The query set was submitted one at a time to PubMed by Agilent Literature Search. The resulting documents were retrieved, parsed into sentences, and analyzed for known interaction terms such as ‘binding’ or ‘activate’. Agilent Literature Search uses a lexicon set to define gene names (concepts) and aliases, drawn from Entrez Gene, and interaction terms (verbs) of interest. An association was extracted from every sentence containing at least two concepts and one verb. Associations were then converted into interactions with corresponding sentences and source hyperlinks, and added to a Cytoscape network [8]. The last download of abstracts was executed on 29 October 2015. In total, 9908 PubMed abstracts were obtained and served as the initial corpus for further processing.
The literature mining method has problems including ‘term variation’ and ‘term ambiguity’ [9]. Term variation originates from the ability of a natural language to express a single concept in a number of ways. For example, in biomedicine, there are many synonyms for proteins, enzymes, and genes. Having six or seven synonyms for a single concept is not unusual in this domain [10]. In the ARN database, we unified a gene as the official gene symbol. Term ambiguity occurs when the same term is used to refer to multiple concepts. For example, the term “fat” can be a noun or an adjective for “obese”. The two terms are often used in biomedical literature. Searching for “fat” in PubMed returned 187888 results. We found that fat was also used to name a gene or as a universal symbol. Therefore, it was necessary to carry out a manual examination of the results of literature mining to delete the wrong results. During this process, we removed most of the 9908 PubMed abstracts, and only 1449 remained.
Information processing and analysis
During the manual annotation and analysis step, information about experimental settings, node classification, function, and adipogenic impact was marked. For each paper in the ARN database, the experimental setting included the experimental procedure and names of cell lines, and the kind of samples was also classified. To store and access the collected information of adipogenesis regulatory networks, we implemented a database and a user-friendly web interface. The ARN database is a Microsoft SQL Server relational database. The table structure of the database is illustrated in Fig. 2, and its complete content is shown in Additional file 1: Table S1.
Table structure of the database. Table “Node” includes the information of the nodes (genes and microRNAs) that regulate adipogenesis. Table “Papers” includes the information of papers about adipogenesis. Table “Summary” includes the summary of the nodes. Table “SNPs” includes information about the SNPs of nodes. Table “Expression” includes the information about expression of nodes in different conditions. Table “Relations” includes the information about the relations of nodes. Table “Material” includes information about the experimental materials in papers. Table “Experiment” includes information about the experiments that were used to verify the relations of nodes
Screening the data of four external databases
Sometimes, the dormant value can only be revealed by combining one dataset with another, perhaps a very different dataset. For example, we obtained 748 transcription factors and their 2347 targets according to 8215 records in transcriptional regulatory relationships unravelled by sentence-based text-mining (TRRUST) database. Next, we concluded that 3538 TF-Target records in TRRUST may be associated with adipogenesis according to the 3053 nodes in the ARN database (Fig. 3). Using the same method, we screened the other three databases. Their basic information is shown in Table 1. In the future, when a new professional database appears, we could rapidly add data associated with adipogenic differentiation to the ARN database according to this method.
Design of the analysis tool
Our interest in text-based scientific discovery led us to the development of the ARN-Analysis tool. Because we envision text-based discovery as a human-centered activity, our goal has been to codify a practical tool that assists a biomedical researcher in formulating and initially testing hypotheses [11].
Generating hypotheses
As shown in Fig. 2 and Additional file 1: Table S1, the information is structured in the ARN database. Therefore, the discovery question is user generated on which subject the user wants to obtain new knowledge. Additionally, the filtering and selection of interesting B- or C-concepts is user dependent. Interesting in this case means interesting according to the current knowledge and goals of the user. It is the user who will have to make an interpretation of the computer-suggested list of possible results. Finally, the intersection of two or more result sets can be obtained by the user, which is likely to be hypotheses.
Testing hypotheses
Once we have many hypotheses, which may be obtained by the ARN-Analysis tool, we may want to determine which is the most important by initially screening out nodes that are highly correlated with adipogenesis. For this purpose, we calculated the “IF” value for each node with the following formula:
In this formula, IF (i) is the effect of node i on the differentiation of fat. Ri is the number of relationships of node i, Rmax is the number of relationships of node r-max that has the most relations; Ei is the number of expression records of node i. Emax is the number of expression records of node e-max that has the most expression records; Pi is the number of prediction records of node i. Pmax is the number of prediction records of node p-max that has the most prediction records. All values are updated with the database, so the information they contain is comprehensive and timely.
Utility
Basic information of the ARN database
Currently, the database contains 3054 nodes (genes and microRNAs), 1807 relation records, 1141 summary records, 10675 expression records, and 43 review images associated with adipogenesis according to 1457 papers. Among the 3054 nodes in the ARN database, we determined 12869 possible relationships sourced from miRGate, TRRUST, BioGRID and PAZAR.
The database can be searched using a web interface (http://210.27.80.93/arn/) [12] with three possible input forms depending on the user’s research focus. For gene searches, Entrez GeneID and official gene symbols are accepted. MicroRNAs require the names of mature microRNA sequences (e.g., mirn143). The literature requires the PubMed PMID (see Additional file 2: Handbook of ARN, Example 1). We provide the node, maps, literature, and expression pages for different kinds of information. Users can select their requested entry and the results page is displayed.
Correlations between databases
A total of 12869 predicted results were obtained by screening the data of four external databases. They are the product of correlations between the ARN database and external databases. As shown in Fig. 4, in the node page, the prediction relations of each node are plotted as a graph, and the relevant information is provided in the form at the same time.
ARN-Analysis is a professional analysis tool for the study of adipogenesis
The analysis page was constructed to screen functional genes and microRNAs based on all the information in the ARN database. Figure 5 shows the three analytical models that we designed for the needs of researchers.
For nodes, researchers can use the “filter by nodes” page, according to the node type, its effects on adipogenesis, to screen node itself, its targets, its source nodes, its predicted source nodes, or its predicted targets. For example, Kim et al. reported that overexpression of mirn21 enhances the differentiation of adipocytes [13]. Therefore, we hypothesized that mirn21 promotes the formation of fat by reducing the expression of proteins that inhibit adipogenesis. Specifically, we needed to discover which mirn21 target genes may inhibit the formation of fat. As shown in Fig. 6, we defined “A” as “adipogenesis”, “B” as “gene”, “C” as “mirn21”, and started from A and C simultaneously, searched for the intersection of the two result sets, and found the answer. The operational procedure was as follows. In the “ARN-Analysis” page, we first entered “mirn21” in the “Symbol” text box. Second, we selected “Prediction Target” in the “Second Screening” options. Third, we clicked on “Filter & Preview”, and there were 60 genes in the result set. We then clicked “Save” to save the “C to B” result set. To obtain the “A to C” result set, we first clicked “Add Result Set” and then selected “Pro-adipogenesis” in the “Differentiation Direction” options. Here, we obtained 173 genes in the result set and then clicked "Save" to save the “A to B” result set. Finally, we clicked “Analysis” and obtained the intersection of the two result sets, which was the answer. It suggested that mirn21 may promote adipogenesis by inhibiting two pro-adipogenesis proteins, NFAT5 and Reck.
Discovery process as defined by Weeber et al [53]. The process is a two-way discovery process that starts from A and C simultaneously, and then finds the intersection B
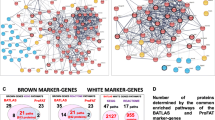
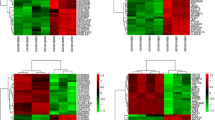
In addition, because there are thousands of differentially expressed nodes in the result sets of many high-throughput articles, usually the author will focus on analysis of a few of them, but most of the remaining nodes may contain important information. Therefore, we designed “Filter By Expression”. Additional file 3: Table S2 contains all of the throughput article PMIDs in the ARN database. Researchers can analyze each of the result sets according to Additional file 3: Table S2. Researchers can also first obtain the throughput article’s PMID in the “ARN-literature page” (see Additional file 2: Handbook of ARN, Example 2), and then use "Filter By Expression" to analyze it. For example, we chose one paper (PMID 25983326) in the “ARN-literature page” found by “advanced search”. This paper describes changes in microRNA expression of white and brown adipose tissues in cold-induced mice [14]. Cold stimulation contributes to the formation of brown fat [15]. Therefore, we hypothesized that some up-regulated microRNAs may inhibit “Anti-browning adipogenesis” proteins, while some down-regulated miRNAs may inhibit “Pro-browning adipogenesis” proteins. Next, we performed the analysis in ARN as shown in Figs. 7 and 8. The results showed that one up-regulated microRNA and 12 down-regulated microRNAs were consistent with our hypothesis. By further analysis in ARN, we obtained the results shown in Table 2.
Furthermore, some researchers need to analyze the result set of their experiment. Therefore, we designed “Enter Symbols”, which allows users to enter a series of genes or microRNAs for analysis. When Xiaoxi et al. performed transcriptome profiling of white adipose tissue in a mouse model for 15q duplication syndrome, they found 145 significantly up-regulated and 85 significantly down-regulated genes [16]. We entered them separately into the ARN-Analysis user-defined input box (see Additional file 2: Handbook of ARN, Example 5). The results of the analysis are shown in Fig. 9: Among the 145 up-regulated genes, 47 nodes were recorded in the ARN database and 98 nodes were newly discovered. Among the 85 down-regulated genes, 30 nodes were recorded in the ARN database and 55 nodes were newly discovered. Combined with other information about the known nodes in the database, we can construct hypotheses, design experiments, and perform further research.
Scoring function of the ARN database
“IF” of the node is obtained from the latest data in the ARN database. When we add a new node or information to the database, the IF of all nodes associated with it will be changed accordingly. This process ensures the timeliness and completeness of the information obtained by researchers. On the home page of the database, we provided the "Link Number" top 50 "hub" nodes. In Table 3, we compared it with the 50 nodes with the maximum “IF” value. Because the IF value indicates the importance of the node to the formation of fat, we assumed that the difference between the two included some new “hub” nodes that may contain certain regulatory relationships that remain to be verified. Using “sp1” as an example, by searching the database node page, we found that expression of the sp1 gene was significantly higher in the liver of cold-induced mice [17]. Therefore, we hypothesized that it is related to brown adipogenesis. Next, we carried out the analysis as shown in Fig. 10. The results suggested that SP1 may regulate the formation of brown fat by regulating “adipoq” [18], “ptges” [19], “rb1”, “sirt1” [20], or “socs1” [21] (see Additional file 2: Handbook of ARN, Example 2). Researchers can design experiments to verify the results. The same method can also be used to analyze the other top 50 nodes.
Screening the intersection between targets of Sp1 and pro-browning adipogenesis genes. The red box indicates the screening condition. See Additional file 2: Handbook of ARN, Example 2 for detailed information
Discussion
Target control of adipogenesis genes
Target control is controlling a subset of target nodes (or a subsystem), which is essential for the system’s mission pertaining to a selected task [22]. Only when we know all the relations of a node can we then know how to control it. The ARN database provides an overview of each node in the adipogenesis regulation network. As shown in Fig. 11 for the node PPAR-gamma, epigenetic modification of its chromatin [23–27], transcriptional regulation of its promoters [28–34], post-transcriptional regulation by microRNAs [35–38], and phosphorylation of its proteins by signaling factors [39, 40] from transcription initiation to the final degradation, this map describes its destiny. This information may help us to design an ideal strategy to control it. Using this information, we may be able to design target control approaches in the future.
Deficiencies of the ARN database
In the process of adding the prediction relations to the ARN database through the external database, we found that the table structure of “Prediction” in Fig. 2 is inadequate. It lacks the relevant tags of the information source database, which obscures the specific sources of the prediction relations. In the future, we will correct this problem by upgrading the platform. In addition, for the IF calculation formula of each node at present, we determined the weights of Ri, Ei and Pi as 1/3. However, with continuous updating and improvement of the platform, the optimal weight of each influencing factor remains to be explored further.
Future directions
The precursors of adipocytes, mesenchymal stem cells (MSCs), can also differentiate into osteoblasts, chondrocytes, and myoblasts. Understanding the factors that govern MSC differentiation has significant implications in diverse areas of human health from obesity to osteoporosis [41]. Therefore, we would like to add them to our network in the future. Moreover, recently, long-chain non-encoding RNA (lncRNA) was found to be involved in the regulation of adipogenic differentiation [42, 43]. These data must be added as soon as they are available. Furthermore, information on the institutions in the papers will soon be added. We are certain that this addition will promote the exchange of ideas, project cooperation, and resource sharing between institutions. We plan to update the database monthly to provide state-of-the-art knowledge and keep track of improvements in the field. All recently added data will be displayed separately on the corresponding page.
Conclusions
The ARN database will serve as a platform for information and hypothesis generation for the research community, which will facilitate uncovering the complexity of adipogenesis-related mechanisms, pathways, and processes.
Availability and requirements
Project name: ARNdbProject. Home page: http://210.27.80.93/arn/. Operating system(s): Platform independent. Other requirements: Microsoft SQL Server,. NET and HTML5 for the Web interface. For interactive data visualization, we applied D3.
Abbreviations
IF, Impact Factor; ARN, Adipogenic Regulation Network; TRRUST, transcriptional regulatory relationships unravelled by sentence-based text-mining; MSCs, mesenchymal stem cells; lncRNA, long-chain non-encoding RNA
References
Hausman DB, DiGirolamo M, Bartness TJ, Hausman GJ, Martin RJ. The biology of white adipocyte proliferation. Obesity Rev. 2001;2:239–54.
Sarjeant K, Stephens JM. Adipogenesis. Cold Spring Harb Perspect Biol. 2012;4(9):a008417.
Garten Y, Coulet A, Altman RB. Recent progress in automatically extracting information from the pharmacogenomic literature. Pharmacogenomics. 2010;11(10):1467–89. doi:10.2217/pgs.10.136.
Swanson DR. 2011 literature-based resurrection of neglected medical discoveries. J Biomed Discov Collab. 2010;6:34–47.
Marc Weeber Klein H, de Jong-van den Berg LTW, Vos R. Using concepts in Literature-Based Discovery: Simulating Swanson’s Raynaud–Fish Oil and Migraine–Magnesium Discoveries. J Am Soc Inf Sci Technol. 2001;52:548–57.
Hur J, Sullivan KA, Schuyler AD, Hong Y, Pande M, States DJ, Jagadish HV, Feldman EL. Literature-based discovery of diabetes- and ROS-related targets. BMC Med Genomics. 2010;3:49.
Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;1211:722–34.
Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;210:2366–82.
Spasic I, Ananiadou S, McNaught J, Kumar A. Text mining and ontologies in biomedicine: making sense of raw text. Brief Bioinform. 2005;63:239–51.
Rojas I, Bernardi L, Ratsch E, Kania R, Wittig U, Saric J. A database system for the analysis of biochemical pathways. In Silico Biol. 2002;2(2):75–86.
Weeber M, Klein H, Aronson AR, Mork JG, de Jong-van den Berg LT, Vos R. Text-based discovery in biomedicine: the architecture of the DAD-system. Proc AMIA Symp. 2000:903-7. PMID:11080015, and PMCID: PMC2243779.
The Adipogenesis Regulation Network database. http://210.27.80.93/arn/. Accessed 15 Jan 2016.
Kim YJ, Hwang SJ, Bae YC, Jung JS. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells. 2009;27(12):3093–102. doi:10.1002/stem.235.
Tao C, Huang S, Wang Y, Wei G, Zhang Y, Qi D, Wang Y, Li K. Changes in white and brown adipose tissue microRNA expression in cold-induced mice. Biochem Biophys Res Commun. 2015;4633:193–9.
Trajkovski M, Lodish H. MicroRNA networks regulate development of brown adipocytes. Trends Endocrinol Metab. 2013;249:442–50.
Liu X, Tamada K, Kishimoto R, Okubo H, Ise S, Ohta H, Ruf S, Nakatani J, Kohno N, Spitz F, et al. Transcriptome profiling of white adipose tissue in a mouse model for 15q duplication syndrome. Genom Data. 2015;5:394–6.
Shore AM, Karamitri A, Kemp P, Speakman JR, Graham NS, Lomax MA. Cold-induced changes in gene expression in brown adipose tissue, white adipose tissue and liver. PLoS One. 2013;87:e68933.
Barth N, Langmann T, Schölmerich J, Schmitz G, Schäffler A. Identification of regulatory elements in the human adipose most abundant gene transcript-1 (apM-1) promoter: role of SP1/SP3 and TNF-alpha as regulatory pathways. Diabetologia. 2002;45(10):1425–33.
Deckmann K, Rörsch F, Steri R, Schubert-Zsilavecz M, Geisslinger G, Grösch S. Dimethylcelecoxib inhibits mPGES-1 promoter activity by influencing EGR1 and NF-kB. Biochem Pharmacol. 2010;809:1365–72.
Li N, Muthusamy S, Liang R, Sarojini H, Wang E. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech Ageing Dev. 2011;1323:75–85.
Madonna S, Scarponi C, Sestito R, Pallotta S, Cavani A, Albanesi C. The IFN-gamma-dependent suppressor of cytokine signaling 1 promoter activity is positively regulated by IFN regulatory factor-1 and Sp1 but repressed by growth factor independence-1b and Krüppel-like factor-4, and it is dysregulated in psoriatic keratinocytes. J Immunol. 2010;185(4):2467–81. doi:10.4049/jimmunol.1001426.
Gao J, Liu YY, D'Souza RM, Barabási AL. Target control of complex networks. Nat Commun. 2014;5:5415.
Jin Q, Wang C, Kuang X, Feng X, Sartorelli V, Ying H, Ge K, Dent SY. Gcn5 and PCAF regulate PPARγ and Prdm16 expression to facilitate brown adipogenesis. Mol Cell Biol. 2014;3419:3746–53.
Cho YW, Hong S, Jin Q, Wang L, Lee JE, Gavrilova O, Ge K. Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis. Cell Metab. 2009;101:27–39.
Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, Rosenbaum M, Zhao Y, Gu W, Farmer SR, Accili D. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell. 2012;1503:620–32.
Chen YH, Yeh FL, Yeh SP, Ma HT, Hung SC, Hung MC, Li LY. Myocyte enhancer factor-2 interacting transcriptional repressor MITR is a switch that promotes osteogenesis and inhibits adipogenesis of mesenchymal stem cells by inactivating peroxisome proliferator-activated receptor gamma-2. J Biol Chem. 2011;28612:10671–80.
Takada I, Kouzmenko AP, Kato S. Wnt and PPARgamma signaling in osteoblastogenesis and adipogenesis. Nat Rev Rheumatol. 2009;58:442–7.
Tong Q, Dalgin G, Xu H, Ting CN, Leiden JM, Hotamisligil GS. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 2000;2905489:134–8.
Banerjee SS, Feinberg MW, Watanabe M, Gray S, Haspel RL, Denkinger DJ, Kawahara R, Hauner H, Jain MK. The Krüppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J Biol Chem. 2003;2784:2581–4.
Mori T, Sakaue H, Iguchi H, Gomi H, Okada Y, Takashima Y, Nakamura K, Nakamura T, Yamauchi T, Kubota N, Kadowaki T, Matsuki Y, Ogawa W, Hiramatsu R, Kasuga M. Role of Krüppel-like factor 15 KLF15 in transcriptional regulation of adipogenesis. J Biol Chem. 2005;28013:12867–75.
Zhao Y, Zhang YD, Zhang YY, Qian SW, Zhang ZC, Li SF, Guo L, Liu Y, Wen B, Lei QY, Tang QQ, Li X. p300-dependent acetylation of activating transcription factor 5 enhances C/EBPβ transactivation of C/EBPαduring 3 T3-L1 differentiation. Mol Cell Biol. 2014;343:315–24.
Pi J, Leung L, Xue P, Wang W, Hou Y, Liu D, Yehuda-Shnaidman E, Lee C, Lau J, Kurtz TW, Chan JY. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J Biol Chem. 2010;28512:9292–300.
Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;4647288:619–23.
Lee H, Kim HJ, Lee YJ, Lee MY, Choi H, Lee H, Kim JW. Krüppel-like factor KLF8 plays a critical role in adipocyte differentiation. PLoS One. 2012;712:e52474.
Zhang JF, Fu WM, He ML, Xie WD, Lv Q, Wan G, Li G, Wang H, Lu G, Hu X, Jiang S, Li JN, Lin MC, Zhang YO, Kung HF. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol. 2011;85:829–38.
Sun J, Wang Y, Li Y, Zhao G. Downregulation of PPARγ by miR-548d-5p suppresses the adipogenic differentiation of human bone marrow mesenchymal stem cells and enhances their osteogenic potential. J Transl Med. 2014;12:168.
Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S, Martindale JL, Hutchison ER, Kim HH, Marasa BS, Selimyan R, Egan JM, Smith SR, Fried SK, Gorospe M. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2011;314:626–38.
Kim SY, Kim AY, Lee HW, Son YH, Lee GY, Lee JW, Lee YS, Kim JB. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun. 2010;3923:323–8.
Hu E, Kim JB, Sarraf P, Spiegelman BM. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARgamma. Science. 1996;2745295:2100–3.
Lemkul JA, Lewis SN, Bassaganya-Riera J, Bevan DR. Phosphorylation of PPARγ Affects the Collective Motions of the PPARγ-RXRα-DNA Complex. PLoS One. 2015;105:e0123984.
Fakhry M, Hamade E, Badran B, Buchet R, Magne D. Molecular mechanisms of mesenchymal stem cell differentiation towards osteoblasts. World J Stem Cells. 2013;5(4):136–48. doi:10.4252/wjsc.v5.i4.136.
Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, Yuan B, Kellis M, Lodish HF, Rinn JL. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;1109:3387–92.
Chen J, Cui X, Shi C, Chen L, Yang L, Pang L, Zhang J, Guo X, Wang J, Ji C. Differential lncRNA expression profiles in brown and white adipose tissues. Mol Genet Genomics. 2015;2902:699–707.
The PubMed Database. http://www.ncbi.nlm.nih.gov/pubmed. Accessed 20 Oct 2015.
The Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining Database. http://www.grnpedia.org/trrust/. Accessed 15 Jan 2016.
Han H, Shim H, Shin D, Shim JE, Ko Y, Shin J, Kim H, Cho A, Kim E, Lee T, et al. TRRUST: a reference database of human transcriptional regulatory interactions. Sci Rep. 2015;5:11432. doi:10.1038/srep11432.
The Public database of Transcription Factor and Regulatory Sequence Annotation. http://www.pazar.info/. Accessed 15 Jan 2016.
Portales-Casamar E, Arenillas D, Lim J, Swanson MI, Jiang S, McCallum A, Kirov S, Wasserman WW. The PAZAR database of gene regulatory information coupled to the ORCA toolkit for the study of regulatory sequences. Nucleic Acids Res. 2009;37(Database issue):D54–60. doi:10.1093/nar/gkn783.
The miRGate database. http://mirgate.bioinfo.cnio.es/miRGate/. Accessed 15 Jan 2016.
Andrés-León E, González Peña D, Gómez-López G, Pisano DG. miRGate: a curated database of human, mouse and rat miRNA-mRNA targets. Database (Oxford). 2015 Apr 8;2015:bav035. doi: 10.1093/database/bav035.
The Biological General Repository for Interaction Datasets. http://thebiogrid.org/. Accessed 15 Jan 2016.
Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–9.
Weeber M, Klein H, de Jong-van den Berg LTW, et al. Using concepts in literature-based discovery: simulating Swanson's Raynaud-fish oil and migraine-magnesium discoveries[J]. J Am Soc Inf Sci Technol. 2001;527:548–57.
Acknowledgements
The authors appreciate the contribution of Mr. Jiang wei-qiang for his help in constructing the database. The authors would also like to thank the generous technical supports from staff of National Beef Cattle Improvement Center for this study.
Funding
This work was supported by the National “86” Program of China (2013AA102505, 2011AA100307-02), the National Science and Technology Support Project of China (2011BAD28B04-03), the GMO New Varieties Major Project of China (2011ZX08007-002), the National Beef and Yak Industrial Technology System of China (CARS-38), the National Natural Science Foundation of China (31272411), and the Scientific and Technological Innovation Program of Shaanxi Province in China (2014KTZB02-02-1).
Availability of data and materials
All the contents in the ARN database are in Additional file 1.
All papers in ARN can be found in http://www.ncbi.nlm.nih.gov/pubmed [44] by entering their PMID.
Project home page: http://210.27.80.93/arn/
Authors’ contributions
LSZ conceived the project and provided final approval of the manuscript version to be published; YH made substantial contributions to conception and design, acquisition of data, and analysis and interpretation of data; YH and LW were involved in drafting the manuscript and revising it critically for important intellectual content. All authors read and approved the final manuscript.
Competing interests
All authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Table S1.
All tables of the ARN database. (XLSX 3574 kb)
Additional file 2:
Handbook of ARN. (PDF 15192 kb)
Additional file 3: Table S2.
All of the throughput articles’ PMID in ARN database. (XLSX 511 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Huang, Y., Wang, L. & Zan, a.Ls. ARN: analysis and prediction by adipogenic professional database. BMC Syst Biol 10, 57 (2016). https://doi.org/10.1186/s12918-016-0321-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12918-016-0321-0