Abstract
Background
The characterization of staphylococcal species that colonize pets is important to maintain animal health and to minimize the risk of transmission to owners. Here, the prevalence of Staphylococcus spp. and methicillin resistance was investigated in canine and feline isolates, and risk factors of staphylococcal colonization were determined. Pets were examined and separated into four groups: (1) healthy dogs, (2) healthy cats, and (3) dogs and (4) cats with clinical signs of bacterial infections of skin, mucous membranes, or wounds. Specimens were collected by a veterinary physician from six anatomic sites (external ear canal, conjunctival sacs, nares, mouth, skin [groin], and anus). In total, 274 animals (cats n = 161, dogs n = 113) were enrolled.
Results
Staphylococcus species were highly diverse (23 species; 3 coagulase-positive and 20 coagulase-negative species), with the highest variety in healthy cats (19 species). The most frequent feline isolates were S. felis and S. epidermidis, while S. pseudintermedius was the most prevalent isolate in dogs. Risk factors of staphylococcal colonization included the presence of other animals in the same household, medical treatment within the last year, and a medical profession of at least one owner. Methicillin resistance was higher in coagulase-negative (17.86%) compared to coagulase-positive (1.95%) staphylococci. The highest prevalence of methicillin-resistant CoNS colonization was observed in animals kept in homes as the most common (dogs and cats).
Conclusions
The association of methicillin-resistant CoNS colonization with animals most often chosen as pets, represents a high risk of transmission between them and owners. The importance of nosocomial transmission of CoNS was also confirmed. This information could guide clinical decisions during the treatment of veterinary bacterial infections. In conclusion, the epidemiologic characteristics of CoNS and their pathogenicity in pets and humans require further research.
Similar content being viewed by others
Background
Staphylococci colonize both living organisms and inanimate surfaces, and can survive adverse environmental conditions. The Staphylococcus genus is comprised of two groups that are categorized by their production of the enzyme coagulase, constituting coagulase-positive (CoPS) and coagulase-negative staphylococci (CoNS) [1]. The greatest staphylococcal threat to human health is posed by the CoPS S. aureus. In dogs, and to a lesser extent in cats, a similar risk is associated with S. pseudintermedius [2, 3]. The most prevalent CoNS species that colonize humans are S. epidermidis, S. haemolyticus, S. capitis, S. hominis, and S. simulans. In comparison, the most common species in pets are S. felis, the S. sciuri group (including S. lentus), S. capitis, and S. cohnii [4, 5]. Until recently, CoNS were considered non-pathogenic or culture-based contaminants; however, this heterogeneous group is now considered clinically significant because of virulence factors and pathogenicity [1, 5,6,7].
Increasing staphylococcal virulence, especially among CoNS, is of concern. The transmission of virulence factors through horizontal gene transfer may confer drug resistance and biofilm production, which confound the treatment of staphylococcal infections [8,9,10,11]. Staphylococci can also survive intracellularly, and thus evade host immune defenses [4, 12, 13].
Both transient and chronic staphylococcal carriage have been described in multiple host species [14,15,16]. Staphylococcal colonization of healthy people and animals occurs at many cutaneous and mucosal sites including ears, conjunctival sacs, nares, mouth, skin, and anus [3, 4, 11, 17,18,19,20]. Staphylococcal infections of animals involve traumatic and surgical wounds; the bloodstream (especially in the presence of infected intravascular devices); the external ear canal; the upper respiratory, urinary, and genital tracts; bone and bone marrow, conjunctiva, and skin [1, 4, 21,22,23,24,25,26]. These infections may feature purulent exudate, tissue necrosis, and even sepsis [27,28,29]. Staphylococcal infections of humans include furunculosis, mastitis, conjunctivitis, keratitis, dacryocystitis, bacteremia, and superantigen-mediated diseases such as scalded skin and toxic shock syndromes [16, 20].
Risk factors of infections due to methicillin-resistant staphylococci (MRS) in dogs and cats include prior hospitalization of the animal, frequent visits to the veterinary office, administration of glucocorticosteroids, and antibiotic treatment [23, 30]. Also, equipment in veterinary clinics can be contaminated with staphylococci [29, 31,32,33].
This study evaluated the prevalence of Staphylococcus spp. in healthy and sick dogs and cats. The predilection of different anatomic sites to staphylococcal colonization was investigated, along with risk factors of colonization. Drug resistance was also explored, with a focus on methicillin resistance.
Results
Study population.
From 2019 to 2021, 274 animals were examined at the Department of Epizootiology and Clinic of Bird and Exotic Animals, Faculty of Veterinary Medicine, Wrocław University of Environmental and Life Sciences, Poland. Cats and dogs were assigned to four groups based on data obtained from clinical examinations and diagnostic interviews of owners. The groups were: healthy cats (n = 120), healthy dogs (n = 64), sick cats (n = 41), and sick dogs (n = 49). Sick animals were diagnosed with at least one of the following conditions: conjunctivitis, upper respiratory tract disease, and skin or wound infection.
Isolation of Staphylococcus spp.
Staphylococcus spp. were isolated from 36.81% (265/720) of samples from healthy cats, 32.75% (94/287) of samples from sick cats, 36.46% (140/384) of samples from healthy dogs, and 34.69% (119/343) of samples from sick dogs. Detailed data on positive sampling are presented in Table 1.
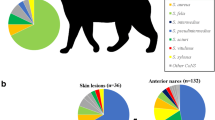
At least one Staphylococcus species was isolated from 81.75% (224/274) of animals. Twenty-three Staphylococcus species were isolated in total. The highest variety of Staphylococcus species was observed in healthy cats (19 species). The most frequently observed species in relation to the number of all staphylococcal isolates obtained from a given group of animals (both healthy and sick) were S. felis (n = 90; 25.07%; Cl 95%: 20.59–29.55%) and S. epidermidis (n = 65; 18.11%; Cl 95%: 14.12–22.09%) in cats, and S. pseudintermedius (n = 138; 53.28%; Cl 95%: 47.21–59.36%) and S. epidermidis (n = 42; 16.22%; Cl 95%: 11.73–20.71%) in dogs.
The prevalence rates of the most frequently isolated staphylococci in each animal group are shown in Fig. 1. Only S. felis in cats and S. pseudintermedius in dogs were isolated from all swabbed anatomic sites. Data on the prevalence of staphylococcal species and their anatomic distributions are presented in Tables 2 and Table 3. The numbers of CoPS and CoNS strains obtained from each group are presented in Table 4.
Detection of methicillin resistance
A total of 597 staphylococcal isolates (96.6% of total collected (597/618)) that had grown after storage at -80 °C were tested for methicillin resistance. From a group of CoNS isolates (from both healthy and sick cats and dogs) (n = 392), 70 isolates were oxacillin-resistant (17.86%; Cl 95%: 14.07–21.65%). In a group of CoPS isolates from both healthy and sick animals (n = 205), only four isolates exhibited oxacillin resistance (1.95%; Cl 95%: 0.06–3.84%). Overall, 22.36% (Cl 95%: 15.92–28.80%) of both healthy and sick cats and 16.81% (Cl 95%: 9.92–23.71%) of dogs were colonized with MRS. The prevalence rates of the most frequently isolated MRS are shown in Fig. 1.
Statistical analysis
A questionnaire regarding risk factors was completed and returned by the owners of 80% of animals. Analysis of these questionnaires identified the presence of other animals in the same household, medical treatment within the last year, and a medical profession of at least one owner as important risk factors of staphylococcal colonization. CoPS colonization was unrelated to medical professions of owners. Breed, age, and sex were unrelated to the isolation of Staphylococcus spp. Animals that lived with other pets in the same household carried more Staphylococcus species (17 in cats, 16 in dogs) compared to those kept individually (11 in cats, 12 in dogs). On the contrary, S. warneri and S. felis were isolated more frequently from individually-kept cats and dogs, respectively (Table 5).
Discussion
This study demonstrated that both healthy and sick pets carry both CoPS and CoNS; however, some species were more highly associated with selected groups of animals. The prevalence of Staphylococcus spp. was nearly 82%, supporting previous studies [34]. Healthy cats exhibited the highest variety of staphylococcal isolates. This result contradicted those obtained by Ma et al. [35], in which dogs carried the most diverse range of Staphylococcus spp. This discrepancy may have been due to the disproportionately large sample size of healthy cats in our study.
Our finding that animals sharing the same households carried more Staphylococcus species suggests transmission between animals living in groups. The higher prevalence of S. warneri and S. felis in individually-kept cats and dogs, respectively, should be examined in further studies, because significantly fewer dogs were tested than cats. This is particularly interesting because S. felis is isolated primarily from cats [4, 5]. Unfortunately, we do not have information regarding previous cat contact of either the S. felis-positive dogs or their owners, and therefore the origin of their strains is unknown. Perhaps S. warneri and S. felis are at a competitive disadvantage against the predominant Staphylococcus species that colonize animals kept in groups, and are therefore more often observed in individually-kept animals.
Interestingly, CoPS were isolated more frequently from dogs (61.4%) compared to cats (12.8%), whereas more CoNS were isolated from cats (87.2%) compared to dogs (38.6%). Our results were similar to those of Abdel-Moein et al. [7] but contrasted with those of Sukur et al. [36]. The high rate of CoPS in dogs is associated with frequent carriage of S. pseudintermedius, as confirmed by our study and previous reports, while in cats both S. aureus and S. pseudintermedius are much less common [2, 3, 26]. This result is important when considering the high methicillin resistance rate of CoNS isolates worldwide [5, 7, 34]. Few studies have compared the virulence factors of CoNS and CoPS [1]; however, both groups are now considered major pathogens, due to drug resistance, possible horizontal gene transfer, and zoonotic potential [1, 4, 5, 7]. The highest methicillin resistance rate was observed among CoNS (nearly 18%) in the current study, in contrast to only 2% in the CoPS group. This result suggests that the pathogenicity of CoNS may be underrated in clinical practice. Becker et al. [4] described CoNS (from both humans and animals) as a reservoir of mobile genetic elements encoding β-lactam- and multidrug resistance. Abdel-Moein et al. [36] suggested that CoNS is a reservoir of resistance genes that can be transmitted to S. aureus, while Argemi et al. [37] showed that CoNS harbored multiple genes responsible for adhesion, biofilm formation, enzyme production, and the encoding of superantigens. These data are of concern, especially in the context of nosocomial infections [4, 5, 36, 37]. Our study showed that the medical profession (both veterinary and human medicine) of the owner, treatment received by the animal over the preceding year, and hospitalization of the owner had a more statistically significant influence on CoNS isolation (S. cohnii, S. epidermidis, S. haemolyticus, S. hominis, S. simulans, S. warneri) compared to CoPS. This shows that exposures to medical facilities, whether due to the owner’s occupation or the patient status of either owner or pet, increases the risk of CoNS colonization. Therefore, medical facilities should be considered as potential reservoirs of these bacteria. Interestingly, such associations were previously ascribed more often to CoPS. Staphylococcus pseudintermedius was isolated more frequently from cats that were treated over the preceding year. In contrast, isolation of S. aureus from the tested animals was not correlated to any factor, contradicting the assumption that it is a most highly pathogenic Staphylococcus species associated with nosocomial infections [38,39,40]. A growing body of evidence indicates that hospitals and veterinary offices are sources of highly pathogenic bacteria, such as CoPS (especially methicillin-resistant S. aureus) [8, 31, 32, 41,42,43]. This study demonstrates that, in fact, not only CoPS (as evidenced by the literature) but also CoNS may be transmitted from medical workplaces to households, and between people and animals. An analysis of colonization of human and pet animal pairs could be an interesting topic for further research.
Interestingly, the presence of children under 12 years of age in the household was a significant risk factor for S. aureus colonization of dogs. This phenomenon might be attributed to children maintaining close physical contact with dogs and having lower hygiene practices. S. aureus colonizing the skin and mucous membranes of children is presumably transmitted to dogs during close physical contact. This confirms that close contact between animals and their owners is a major risk factor for staphylococcal colonization, with both anthroponotic and zoonotic transmission being possible [42, 44,45,46,47].
S. pseudintermedius is isolated frequently from dogs globally [17, 28, 47]. Of note, in the current study, S. pseudintermedius was isolated more frequently from dogs that were in contact with other animals. Unexpectedly, outdoor cats were more likely to be colonized with S. xylosus compared to indoor-only cats; thus, the microbiota of these groups might differ, supporting the findings of Older et al. [48]. This finding might be explained by increased opportunities of outdoor cats for contact with other animals, people, and the external environment.
Despite investigating a large number of samples the current study did have some limitations. First, due to the disproportionately large sample size of healthy cats, it is difficult to accurately compare the prevalence of staphylococci species in other groups of animals. The low values may be due to the relatively small number of animals tested. Moreover, due to the very large total number of obtained Staphylococcus spp. strains, resistance testing at the genotypic level (by PCR) has not been performed. According to the Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals, supplement VET01S, for other Staphylococcus strains (excluding S. aureus, S. lugdunensis, S. epidermidis, S. pseudintermedius and S. schleiferi) oxacillin MIC breakpoints may overcall resistance, and some isolates for which the oxacillin MICs are 0.5–2 μg/mL may be mecA negative. These isolates could be tested for presence of mecA gene or for PBP2a, and if found negative, should be reported as methicillin (oxacillin) susceptible [49]. In this study, these additional methods were not performed. Finally, only oxacillin was used in the MIC resistance test. Considering the above, further research projects should be extended with these methods to obtain more complete results.
Conclusions
This study confirmed that staphylococcal colonization is common in domesticated dogs and cats, with the most prevalent species being S. pseudintermedius in dogs and S. felis and S. epidermidis in cats. CoPS and CoNS were more often isolated from dogs and cats, respectively. Of importance, methicillin resistance was more prevalent in CoNS than in CoPS. The highest rate of methicillin resistance in CoNS isolates was observed in pets kept in homes, suggesting a high risk of both anthroponotic and zoonotic transmission. Consequently, physicians and veterinarians should educate their patients and pet owners on maintaining appropriate hygiene during contact with animals. The importance of nosocomial transmission of CoNS was also confirmed. This information could guide clinical decisions when veterinary patients are treated for bacterial infections. Particular attention should be paid to the role of CoNS in infections of the skin and mucous membranes and to the conduction of a thorough diagnostic evaluation (including bacteriological culture and antibiotic susceptibility testing) and rational drug selection. Unfortunately, CoNS are still considered by some clinicians as non-pathogenic commensals; therefore, their isolation from clinical specimens may not always lead to further analysis, antibiotic susceptibility testing, or targeted antibiotic therapy; resulting in adverse clinical outcomes and the spread of resistance and virulence factors. In conclusion, the epidemiologic characteristics of CoNS and their pathogenicity in pets and humans require further research.
Materials and methods
Study population and sampling procedures
This study extended the animal groups and number of collection sites used in a previous study [50], to obtain more details on staphylococcal colonization of pets. Pets were separated into four groups: (1) healthy dogs, (2) healthy cats, and (3) dogs and (4) cats with clinical signs of bacterial infections on the skin, mucous membranes, or wounds. Animals were only included after receiving permission from the owners to collect samples. Each owner was asked to complete a survey about the pet and household (home environment).
The research project was submitted to the Local Ethics Committee for Animal Experiments in Wrocław, Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Sciences, Poland. Due to the project’s proposed noninvasive sampling methods, the Ethics Committee qualified the study as research, thus exempting it from any further approval from the Ethics Committee. All methods described were approved by Wroclaw University of Environmental and Life Sciences, and were performed in compliance with the relevant guidelines and regulations for good laboratory practices. Each pet owner submitted informed consent to participate in this study, and completed the proper documentation.
Specimens were collected from six anatomic sites of each pet by a veterinary physician at study admission (immediately after obtaining consent of the animal owner for its study participation); these included the external ear canal, conjunctival sacs, nares, oral cavity, skin (groin), and anus. In the groups of sick animals, an extra swab was collected from the diseased wound or skin, if present.
Isolation and identification of Staphylococcus spp. from samples
The collected material was placed in 2 ml of liquid brain–heart infusion broth (BHI) (Oxoid, Basingstoke, United Kingdom). The samples were then incubated at 37 °C for 24 h and then submitted for species identification. Staphylococcus spp. were isolated and identified from samples following an existing method [50]. One microliter of bacterial BHI stock was sub-cultured in mannitol-salt agar and Columbia blood agar plate (Oxoid, Basingstoke, United Kingdom). The plates were then incubated for 24 h at 37 °C. If the culture result was uncertain (only single visible colonies) or negative (no bacterial growth on the plate), incubation was extended to 48 h. After this time, the absence of bacterial growth was considered a negative culture result. Preliminary identification of staphylococci was based on colony morphology on Columbia blood agar and mannitol salt agar simultaneously. Bacterial colonies which grew on Columbia blood agar, diameter 2–6 mm, round, smooth, glistening, opaque, white or more or less yellow pigmented (grey-yellow, yellow-orange, gold), with or without beta-hemolysis zones were selected for subsequent identification. On the mannitol-salt agar plates, yellow colonies with surrounding yellow medium were suspected as CoPS and red colonies without medium color change were suspected as CoNS. The colonies used for the identification were obtained after comparing the growth of bacteria simultaneously on both the used media in order to minimize the risk of making a mistake in the diagnosis. The preliminary identification of suspected strains included Gram staining and detection of enzyme production (coagulase tube test; IBSS Biomed, Cracow, Poland). In case of morphologically distinguishable staphylococcal colonies were visible (maximum 5 colonies), these were cultured separately again in solid medium (Columbia blood agar) to obtain pure colonies.
A single colony from selected, and pure strains were further identified by matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS), following the method of Król et al. [51]. Raw spectra were processed using MALDI Biotyper OC v.3.1. software (Bruker Daltonik GmbH, Bremen, Germany). The results were classified to the species level using score values proposed by the manufacturer. The following identification scores were used: < 1.7 = no reliable identification; 1.7–1.999 = probable identification to the genus level; 2.0–2.299 = secure genus identification, probable species identification; and 2.3–3.0 = highly probable species identification. Scores ≥ 2.0 were considered as acceptable species-level identification [51]. All species were assigned after obtaining a score higher than 2.0. The indicated strains were stored for further analysis in 1 ml of bacterial stock in BHI with 15% glycerol at -80 °C for up to 12 months.
Detection of methicillin resistance
All staphylococcal isolates that had grown after storage in -80 °C were tested for methicillin resistance. Methicillin resistance was detected using oxacillin with the broth microdilution method, using polystyrene, sterile titer plates (FL MEDICAL, Torreglia (PD), Italy). The examined strains were inoculated in cation-adjusted Mueller–Hinton broth (Oxoid, Basingstoke, United Kingdom) and in a dilution series of 4, 2, 1, 0.5, 0.25, and 0.125 µg/mL oxacillin stock solutions (TOKU-E, Gent, Belgium) in water [49, 52]. Inoculum (colony suspension, equivalent to a 0.5 McFarland Standard) was prepared according to the standard broth microdilution procedure described in Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals, supplement VET01S [49]. Samples were incubated for 24 h at 37 °C. Methicillin-resistant strains were detected using CLSI criteria for each Staphylococcus species [49]. S. aureus ATCC 29213 was used as a negative control, and S. aureus ATCC 43300 (mecA-positive) was used as a positive control.
Statistical analysis
Statistical analysis of potential risk factors of colonization by specific Staphylococcus spp. was based on information provided by owners. Animals were excluded if owners did not return or complete the questionnaire. Data on the characteristics of pets, along with their medical history and environmental living conditions, were compared with scores on the frequency of Staphylococcus spp. isolation and oxacillin resistance. Statistical analysis was conducted using the R statistical package (v 3.6.3.). The prevalence and confidence intervals of staphylococci and MRS were calculated using the bootstrap method. All data were analyzed using the Shapiro–Wilk test, Wilcoxon test, Kruskal–Wallis test, Chi-square tests, and Fisher's test. P < 0.05 was considered statistically significant.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Abbreviations
- CoPS:
-
Coagulase-positive staphylococci
- CoNS:
-
Coagulase-negative staphylococci
- BHI:
-
Brain–heart infusion broth
- MALDI-TOF MS:
-
Matrix-assisted laser desorption ionization-time of flight mass spectrometry
- CLSI:
-
Clinical and Laboratory Standards Institute
- MRS:
-
Methicillin-resistant staphylococci
References
França A, Gaio V, Lopes N, Melo LDR. Virulence factors in coagulase-negative staphylococci. Pathogens. 2021;10(2):170. https://doi.org/10.3390/pathogens10020170.
Kizerwetter-Świda M, Chrobak-Chmiel D, Rzewuska M. Problemy weterynaryjnej diagnostyki mikrobiologicznej dotyczącej zakażeń gronkowcowych. Życie Wet. 2018;93:373–6.
van Duijkeren E, Catry B, Greko C, Moreno MA, Pomba MC, Pyörälä S, et al. Review on methicillin-resistant Staphylococcus pseudintermedius. J Antimicrob Chemother. 2011;66(12):2705–14. https://doi.org/10.1093/jac/dkr367.
Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27(4):870–926. https://doi.org/10.1128/CMR.00109-13.
Elnageh HR, Hiblu MA, Abbassi MS, Abouzeed YM, Ahmed MO. Prevalence and antimicrobial resistance of Staphylococcus species isolated from cats and dogs. Open Vet J. 2021;10(4):452–6. https://doi.org/10.4314/ovj.v10i4.13.
Goetz C, Tremblay YDN, Lamarche D, et al. Coagulase-negative staphylococci species affect biofilm formation of other coagulase-negative and coagulase-positive staphylococci. J Dairy Sci. 2017;100(8):6454–64. https://doi.org/10.3168/jds.2017-12629.
Sukur H, Esendal O. Presence and antimicrobial resistance of coagulase-negative staphylococci isolated from animals in a Veterinary Teaching Hospital in Cyprus. Vet Med. 2020;65:191–8.
Paul NC, Moodley A, Ghibaudo G, Guardabassi L. Carriage of methicillin-resistant Staphylococcus pseudintermedius in small animal veterinarians: indirect evidence of zoonotic transmission. Zoonoses Public Health. 2011;58(8):533–9. https://doi.org/10.1111/j.1863-2378.2011.01398.x.
Rodrigues AC, Belas A, Marques C, Cruz L, Gama LT, Pomba C. Risk factors for nasal colonization by methicillin-resistant staphylococci in healthy humans in professional daily contact with companion animals in Portugal. Microb Drug Resist. 2018;24(4):434–46. https://doi.org/10.1089/mdr.2017.0063.
Silva MB, Ferreira FA, Garcia LN, Silva-Carvalho MC, Botelho LA, Figueiredo AM, Vieira-da-Motta O. An evaluation of matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the identification of Staphylococcus pseudintermedius isolates from canine infections. J Vet Diagn Invest. 2015;27(2):231–5. https://doi.org/10.1177/1040638715573297.
Gómez-Sanz E, Ceballos S, Ruiz-Ripa L, Zarazaga M, Torres C. Clonally diverse methicillin and multidrug resistant coagulase negative staphylococci are ubiquitous and pose transfer ability between pets and their owners. Front Microbiol. 2019;10:485. https://doi.org/10.3389/fmicb.2019.00485.
Szabados F, Kleine B, Anders A, Kaase M, Sakınç T, Schmitz I, Gatermann S. Staphylococcus saprophyticus ATCC 15305 is internalized into human urinary bladder carcinoma cell line 5637. FEMS Microbiol Lett. 2008;285(2):163–9. https://doi.org/10.1111/j.1574-6968.2008.01218.x.
Josse J, Laurent F, Diot A. Staphylococcal adhesion and host cell invasion: fibronectin-binding and other mechanisms. Front Microbiol. 2017;8:2433. https://doi.org/10.3389/fmicb.2017.02433.
Hartmann FA, White DG, West SE, Walker RD, Deboer DJ. Molecular characterization of Staphylococcus intermedius carriage by healthy dogs and comparison of antimicrobial susceptibility patterns to isolates from dogs with pyoderma. Vet Microbiol. 2005;108(1–2):119–31. https://doi.org/10.1016/j.vetmic.2005.03.006.
Sollid JU, Furberg AS, Hanssen AM, Johannessen M. Staphylococcus aureus: determinants of human carriage. Infect Genet Evol. 2014;21:531–41. https://doi.org/10.1016/j.meegid.2013.03.020.
Shinefield HR, Ruff NL. Staphylococcal infections: a historical perspective. Infect Dis Clin North Am. 2009;23(1):1–15. https://doi.org/10.1016/j.idc.2008.10.007.
Kuan EC, Yoon AJ, Vijayan T, Humphries RM, Suh JD. Canine Staphylococcus pseudintermedius sinonasal infection in human hosts. Int Forum Allergy Rhinol. 2016;6(7):710–5. https://doi.org/10.1002/alr.21732.
Robb AR, Wright ED, Foster AME, Walker R, Malone C. Skin infection caused by a novel strain of Staphylococcus pseudintermedius in a Siberian husky dog owner. JMM Case Rep. 2017;4(3):jmmcr005087. https://doi.org/10.1099/jmmcr.0.005087.
Somayaji R, Priyantha MA, Rubin JE, Church D. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: report of 24 cases. Diagn Microbiol Infect Dis. 2016;85(4):471–6. https://doi.org/10.1016/j.diagmicrobio.2016.05.008.
Deguchi H, Kitazawa K, Kayukawa K, Kondoh E, Fukumoto A, Yamasaki T, et al. The trend of resistance to antibiotics for ocular infection of Staphylococcus aureus, coagulase-negative staphylococci, and Corynebacterium compared with 10-years previous: a retrospective observational study. PLoS One. 2018;13(9):e0203705. https://doi.org/10.1371/journal.pone.0203705.
Chrobak D, Kizerwetter-Świda M, Rzewuska M, Binek M. Staphylococcus pseudintermedius – nowy, ale dobrze znany patogen. Życie Wet. 2013;88:625–8.
Kalhoro DH, Kalhoro MS, Mangi MH, Jahejo AR, Kumbhar S, Lochi GM, et al. Antimicrobial resistance of staphylococci and streptococci isolated from dogs. Trop Biomed. 2019;36(2):468–74.
Krapf M, Müller E, Reissig A, Slickers P, Braun SD, Müller E, Ehricht R, Monecke S. Molecular characterisation of methicillin-resistant Staphylococcus pseudintermedius from dogs and the description of their SCCmec elements. Vet Microbiol. 2019;233:196–203. https://doi.org/10.1016/j.vetmic.2019.04.002.
Maluping RP, Paul NC, Moodley A. Antimicrobial susceptibility of methicillin-resistant Staphylococcus pseudintermedius isolated from veterinary clinical cases in the UK. Br J Biomed Sci. 2014;71(2):55–7. https://doi.org/10.1080/09674845.2014.11669965.
Marques C, Belas A, Franco A, Aboim C, Gama LT, Pomba C. Increase in antimicrobial resistance and emergence of major international high-risk clonal lineages in dogs and cats with urinary tract infection: 16 year retrospective study. J Antimicrob Chemother. 2018;73(2):377–84. https://doi.org/10.1093/jac/dkx401.
Marszalik A, Chrobak-Chmiel D, Golke A, Sałamaszyńska-Guz A, Dembele K. Staphylococcus pseudintermedius–czy wiemy o nim wszystko? Życie Wet. 2018;93:238–40.
Maali Y, Badiou C, Martins-Simões P, Hodille E, Bes M, Vandenesch F, et al. Understanding the virulence of staphylococcus pseudintermedius: a major role of pore-forming toxins. Front Cell Infect Microbiol. 2018;8:221.
Corrò M, Skarin J, Börjesson S, Rota A. Occurrence and characterization of methicillin-resistant Staphylococcus pseudintermedius in successive parturitions of bitches and their puppies in two kennels in Italy. BMC Vet Res. 2018;14(1):308. https://doi.org/10.1186/s12917-018-1612-z.
Youn JH, Park YH, Hang’ombe B, Sugimoto C. Prevalence and characterization of Staphylococcus aureus and Staphylococcus pseudintermedius isolated from companion animals and environment in the veterinary teaching hospital in Zambia. Africa Comp Immunol Microbiol Infect Dis. 2014;37(2):123–30. https://doi.org/10.1016/j.cimid.2014.01.003.
Lehner G, Linek M, Bond R, Lloyd DH, Prenger-Berninghoff E, Thom N, et al. Case-control risk factor study of methicillin-resistant Staphylococcus pseudintermedius (MRSP) infection in dogs and cats in Germany. Vet Microbiol. 2014;168(1):154–60. https://doi.org/10.1016/j.vetmic.2013.10.023.
Espadale E, Pinchbeck G, Williams NJ, Timofte D, McIntyre KM, Schmidt VM. Are the hands of veterinary staff a reservoir for antimicrobial-resistant bacteria? A randomized study to evaluate two hand hygiene rubs in a veterinary hospital. Microb Drug Resist. 2018;24(10):1607–16. https://doi.org/10.1089/mdr.2018.0183.
Feßler AT, Schuenemann R, Kadlec K, Hensel V, Brombach J, Murugaiyan J, et al. Methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant Staphylococcus pseudintermedius (MRSP) among employees and in the environment of a small animal hospital. Vet Microbiol. 2018;221:153–8. https://doi.org/10.1016/j.vetmic.2018.06.001.
Paul NC. MRSP: prevalence in practice. Vet Rec. 2015;176:170–1.
Ruzauskas M, Couto N, Siugzdiniene R, Klimienė I, Virgailis M, Vaskeviciute L, et al. Methicillin-resistant coagulase-negative Staphylococcus spp. prevalence in lithuanian dogs: a cross-sectional study. Veterinarski Arhiv. 2015;85:175–87.
Ma GC, Worthing KA, Ward MP, Norris JM. Commensal staphylococci including methicillin-resistant staphylococcus Aureus from dogs and cats in remote new south wales. Australia Microb Ecol. 2020;79(1):164–74. https://doi.org/10.1007/s00248-019-01382-y.
Abdel-Moein KA, Zaher HM. The nasal carriage of coagulase-negative staphylococci among animals and its public health implication. Vector Borne Zoonotic Dis. 2020;20(12):897–902. https://doi.org/10.1089/vbz.2020.2656.
Argemi X, Hansmann Y, Prola K, Prévost G. Coagulase-negative staphylococci pathogenomics. Int J Mol Sci. 2019;20(5):1215. https://doi.org/10.3390/ijms20051215.
Churak A, Poolkhet C, Tamura Y, Sato T, Fukuda A, Thongratsakul S. Evaluation of nosocomial infections through contact patterns in a small animal hospital using social network analysis and genotyping techniques. Sci Rep. 2021;18,11(1):1647. https://doi.org/10.1038/s41598-021-81301-9.
Weese JS, Dick H, Willey BM, McGeer A, Kreiswirth BN, Innis B, Leow DE. Suspected transmission of methicillin-resistant Staphylococcus aureus between domestic pets and humans in veterinary clinics and in the household. Vet Microbiol. 2006;115(1–3):148–55. https://doi.org/10.1016/j.vetmic.2006.01.004.
Kisani AI, Awasum A, Udegbunam S, Nnaji T, Muhammed B, Melekwa G, et al. Management of nosocomial diseases in small animal practice: a review. Vom Journal of Veterinary Science. 2016;11:94–100.
Worthing KA, Brown J, Gerber L, Trott DJ, Abraham S, Norris JM. Methicillin-resistant staphylococci amongst veterinary personnel, personnel-owned pets, patients and the hospital environment of two small animal veterinary hospitals. Vet Microbiol. 2018;223:79–85. https://doi.org/10.1016/j.vetmic.2018.07.021.
van Duijkeren E, Kamphuis M, van der Mije IC, Laarhoven LM, Duim B, et al. Transmission of methicillin-resistant Staphylococcus pseudintermedius between infected dogs and cats and contact pets, humans and the environment in households and veterinary clinics. Vet Microbiol. 2011;150(3–4):338–43. https://doi.org/10.1016/j.vetmic.2011.02.012.
Tabatabaei S, Najafifar A, AskariBadouei M, ZahraeiSalehi T, AshrafiTamai I, et al. Genetic characterisation of methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in pets and veterinary personnel in Iran: new insights into emerging methicillin-resistant S. pseudintermedius (MRSP). J Glob Antimicrob Resist. 2019;16:6–10. https://doi.org/10.1016/j.jgar.2018.08.022.
Bierowiec K, Płoneczka-Janeczko K, Rypuła K. Is the colonisation of staphylococcus aureus in pets associated with their close contact with owners? PLoS One. 2016;11(5):e0156052. https://doi.org/10.1371/journal.pone.0156052.
Lozano C, Rezusta A, Ferrer I, Pérez-Laguna V, Zarazaga M, Ruiz-Ripa L, et al. Staphylococcus pseudintermedius human infection cases in Spain: dog-to-human transmission. Vector Borne Zoonotic Dis. 2017;17(4):268–70. https://doi.org/10.1089/vbz.2016.2048.
Kwaszewska A, Lisiecki P, Szemraj M, Szewczyk EM. Zwierzęcy Staphylococcus felis o potencjale zakażania skóry człowieka [Animal Staphylococcus felis with the potential to infect human skin]. Med Dosw Mikrobiol. 2015;67(2):69–78.
Ference EH, Danielian A, Kim HW, Yoo F, Kuan EC, Suh JD. Zoonotic Staphylococcus pseudintermedius sinonasal infections: risk factors and resistance patterns. Int Forum Allergy Rhinol. 2019;9(7):724–9. https://doi.org/10.1002/alr.22329.
Older CE, Diesel AB, Lawhon SD, Queiroz CRR, Henker LC, Rodrigues Hoffmann A. The feline cutaneous and oral microbiota are influenced by breed and environment. PLoS One. 2019;14(7):e0220463. https://doi.org/10.1371/journal.pone.0220463.
CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. 5th ed. CLSI supplement VET01S. USA: Clinical and Laboratory Standards Institute; 2020.
Bierowiec K, Korzeniowska-Kowal A, Wzorek A, Rypuła K, Gamian A. Prevalence of staphylococcus species colonization in healthy and sick cats. Biomed Res Int. 2019;2019:4360525. https://doi.org/10.1155/2019/4360525.
Król J, Wanecka A, Twardoń J, Mrowiec J, Dropińska A, Bania J, et al. Isolation of Staphylococcus microti from milk of dairy cows with mastitis. Vet Microbiol. 2016;182:163–9. https://doi.org/10.1016/j.vetmic.2015.11.018.
Clinical and laboratory standards institute. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—ninth edition. 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087, USA: CLSI document M07-A9, Clinical and Laboratory Standards Institute; 2012.
Acknowledgements
We are indebted to Mr. Grzegorz Golonka for substantive help with data analyzing.
Funding
This work was supported by the Wrocław University of Environmental and Life Sciences (Poland) as the Ph.D. research program "Bon doktoranta SD UPWr” and by the project "ProHum - Interdisciplinary Doctoral School —planning experimental research, creating and optimizing experimental animal models with the ability to transfer them to clinical trials in human medicine” co-financed by the European Social Fund under the Operational Program Knowledge Education Development, under contract POWR.03.02.00–00-I008/17. The APC is financed by Wroclaw University of Environmental and Life Sciences.
DisclosureThe funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
MM and KB have made substantial contributions to the conception of the research; MM collected the material from examined animals, conducted the laboratory work, analyzed the obtained data and was the major contributor in writing the manuscript; AKK, AW and AG made species identification using MALDI-TOF MS method; KB took part in laboratory analyses including bacteriological examination and antimicrobial resistance testing, interpreted the data and participated in the writing of the manuscript; KR and KB supervised the writing of the manuscript and corrected the text. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The research project was submitted to the 2nd Local Ethics Committee for Animal Experiments in Wrocław, Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Sciences, Poland. Due to the noninvasive samples collection procedure, the Ethics Committee qualified the study as research, which did not require any further approval from the Ethics Committee (opinion No 814/2019). All methods described were approved by Wroclaw University of Environmental and Life Sciences and were performed in compliance with the relevant guidelines and regulations for good laboratory practice. Each pet owner informed them that they consented to participate in this study and filled out the proper documentation.
The study is reported in accordance with ARRIVE guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Miszczak, M., Korzeniowska-Kowal, A., Wzorek, A. et al. Colonization of methicillin-resistant Staphylococcus species in healthy and sick pets: prevalence and risk factors. BMC Vet Res 19, 85 (2023). https://doi.org/10.1186/s12917-023-03640-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-023-03640-1