Abstract
Background
Salmonella as an important food-borne zoonotic bacterial pathogen, infection in ducks is a recessive infection, however, it can also cause high mortality and threat to food safety. Preventing and controlling the infection and transmission of Salmonella in ducks critically require rapid and sensitive detection method. Full-length Salmonella-specific protein PagN was induced and expressed in E.coil BL21 and was purified as an antigen to establish an indirect enzyme-linked immunosorbent assays (iELSA) detection kit.
Results
The recombinant PagN protein has a molecular weight of 43 kDa containing a His-tag, was recognized by an anti-Salmonella positive serum by Western blot assay. The optimal concentration of PagN as a coating antigen in the iELISA was 1 μg/mL, and the optimal dilution of enzyme-labeled secondary antibody was 1:4000 (0.025 μg/mL). The cutoff OD450 value was established at 0.268. The iELISA kit showed high selectivity since no cross-reaction with E. coli, Staphylococcus aureus and Streptococcus was observed. iELISA method and Dot-blot test were performed on 100 clinical sera samples collected from duck farms, and the actual coincidence rate was 89% (89/100). 613 duck serum samples from 3 different farms were tested using established method and commercial ELISA kit. The concordance between the two methods was 94.1%.
Conclusion
Anti-PagN based iELISA can serve as a useful tool for diagnosis of Salmonella infection.
Similar content being viewed by others
Background
Salmonellosis in ducks is a common and multiple infectious disease caused by different Salmonella serotypes, which seriously endangers the survival of ducklings. In particular, Salmonella co-infected with Riemerella anatipestifer and E.coli cause enormous morbidity and mortality [1]. Salmonella can be transmitted both horizontally and vertically [2], and lead to a large outbreak of Salmonella infection in a short time if it is not detected in time. In recent years, more and more multi-drug-resistant bacteria have appeared in China, which makes salmonellosis in ducks more difficult to prevent and control [3, 4]. At the same time, Salmonella is one of the most important foodborne pathogens worldwide. Duck meat and other by-products contaminated by Salmonella remains a potential source of human salmonellosis [5, 6]. Salmonella is 1 of the 4 key global causes of diarrhoeal diseases according to the World Health Organization (WHO) guidelines [7]. In the United States, around 20% of foodborne illnesses attributed to Salmonella can be associated with poultry and poultry products [8]. Moreover, the proportion is likely to be higher in developing countries. As the country with the largest duck-raising industry in the world, China needs to strengthen the detection and control of Salmonella. Therefore, the establishment of accurate and rapid diagnostic method is an important way to solve this problem.
Although detection methods relying on traditional bacterial culturing have been used as the gold standard for the detection of Salmonella [9], it is a time-consuming and laborious process, and cannot meet the urgent need of real-time detection. Some Salmonella detection methods have been developed such as: polymerase chain reaction (PCR) [10], quantitative real-time PCR (qPCR) [11, 12], loop-mediated isothermal amplification (LAMP) [13, 14], gold immunochromatographic test strip (ICTS) [15], enzyme-linked immunosorbent assays [16, 17], and biosensors [18, 19]. Among the above methods, conventional iELISA established based on the excellent antigenic protein relevant to high conservation from different serotypes and antigenic specificity is very suitable for rapidly monitoring and purifying the Salmonella contamination in large-scale duck industry in real time.
PagN is an outer membrane protein that contributes to adhesion to and invasion of epithelial cells by Salmonella. The PagN as conserved gene is localized on the specific centisome 7 genomic island and widely distributed among the different Salmonella serovars [20, 21]. The PagN is a PhoP-regulated gene that is up-regulated during growth within macrophages and in vivo in murine models of infection [22]. Some studies have shown that the PagN protein utilizes heparinated proteoglycans to adhere and invade mammalian cells [21, 23]. The PagN protein contains a transmembrane region and four extracellular loops. The latter is very important to the invasion of epithelial cells and can induce a strong immune response [20, 24]. After bioinformatics analysis, we found that there are many potential antigen sites for PagN protein, and the immunogenicity, hydrophilicity, flexibility and surface possibility are relatively good as antigenic determinants. Therefore, PagN could be a promising target for detecting Salmonella antibodies.
In this study, the PagN gene was cloned into the prokaryotic expression vector pET-32a(+) to construct a recombinant expression plasmid and expressed in E. coli BL 21 cells. An iELISA was developed based on purified recombinant PagN for detecting Salmonella antibodies in duck serum. This method has the potential to contribute to effective diagnosis of Salmonellosis in ducks and to gradually eliminate Salmonella infections.
Results
Serum preparation
We found that different positive sera and corresponding bacteria produced agglutination reaction. The liquid in the test tube was clear macroscopically, but the clumps were visible to the naked eye when a hanging drop was examined. More precipitate collected in the bottom of the tubes. On the contrary, there was no agglutination in each negative control group, and the liquid was turbid. The results showed that all the positive and negative sera were obtained, which can be used for the negative control, and sensitivity and specificity detection of iELISA.
Screening of target protein
The homology of PagN gene among Salmonella reached more than 98%, and the homology with other non-Salmonella was less than 50% by BLASTN comparison (Fig. S1). DNAstar software and IEDB website predicted that the PagN protein (43 kDa) had a relatively large proportion of β-turn and random coil, and contained good hydrophilicity, flexibility, immunogenicity and surface possibility (Fig. S2). B-cell epitope prediction showed that four antigenic epitopes with strong antigenicity nearly distributed in the extracellular loop region of the protein, which could stimulate the body to produce specific antibodies for serological detection (Fig. S3).
Recombinant plasmid construction and the recombinant PagN protein with his-tag purification
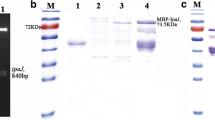
A 651 bp of the PagN gene sequence was successfully amplified by PCR from the Salmonella enteritis strain (CVCC 3377), which was subsequently inserted into the pET-32a(+) vector. The recombinant pET-32a (+)-PagN plasmid was identified by DNA sequencing and double digestion with BamH I and Hind III (Fig. 1). The plasmid was transformed into E.coli BL21 (DE3) cells. The recombinant PagN protein with N-terminal His-tag was induced for 6 h at 37 °C by adding 1 mM isopropyl-b-D-thiogalactopyranoside (IPTG). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis showed that a target band was found at about 43 kDa (lane 5 in Fig. 2A), but no band could be detected for empty vector (lane 3 in Fig. 2A), indicating that recombinant PagN protein was successfully induced. Supernatant and inclusion body samples obtained after ultrasonication in an ice bath were processed, and then subjected to SDS-PAGE analysis showed that the recombinant PagN protein mainly expressed in the form of inclusion bodies (Fig. 2B). Ni2+ NTA agarose gel affinity chromatography was applied to purify the His-tagged recombinant PagN protein, and a single band was detected by SDS-PAGE (Fig. 2C). For western-blot analysis, the recombinant protein can be recognized by duck anti-Salmonella serum (Fig. 2D).
Recombinant PagN protein expression, purification and identification. (A) Induction of expression of the PagN protein. M: Protein molecular weight marker; Lane 1: E.coil BL21(DE3) cells; Lane 2: empty vector pET-32a(+) plasmid transferred into E.coil BL21(DE3) was induced expression without IPTG; Lane 3: empty vector pET-32a(+) plasmid transferred into E.coil BL21(DE3) was induced expression by IPTG; Lane 4: PagN recombinant protein expression without IPTG induction; Lane 5: PagN recombinant protein expression by IPTG induction at 37 °C for 6 h. (B) Analysis of recombinant PagN protein expression patterns. M: Protein molecular weight marker; Lane 1: expression of the target protein in supernatant; Lane 2: expression of the target protein in inclusion bodies. (C) Analysis of the purified recombinant PagN protein by affinity chromatography. M: Protein molecular weight marker; Lane 1: purified PagN protein using nickel affinity chromatography. (D) Western blotting analysis of recombinant PagN protein. M: Protein molecular weight marker; Lane 1, 2: recombinant PagN protein (43 kDa) identified via Western blotting
Establishment of the recombinant PagN protein-based iELISA
The optimal coating antigen concentration as determined by checkerboard titration was 1 μg/mL, and the optimal serum dilution was identified as 1:50 (Table 1). The subsequent reaction conditions as following: the concentration of coating antigen was for 2 h at 37 °C; 5% skimmed milk powder blocked for 2 h at 37 °C; the best serum dilution was 1:50, the incubation condition of serum was at 37 °C for 30 min; the enzyme-labeled secondary antibody at a working dilution of 1:4000 was incubated 90 min at 37 °C, and the catalytic reaction time of tetramethylbenzidine (TMB) was 20 min (Table 2).
Cut-off value determination of the iELISA
The OD450 values of 38 Salmonella negative sera were determined by the established iELISA method. According to the cut-off value = mean value of negative sera + 3 × standard deviation, the cutoff value was determined to be 0.268 (Fig. 3).
Sensitivity of the iELISA
The Salmonella positive sera were continuously diluted by two-fold (1,50 to 112,800) and detected by iELISA. The maximum dilution was 1:1600 according to the established iELISA method (Fig. 4).
Specific determination of the iELISA
The serum cross-reactivity test results showed that the OD450 values of all 3 Salmonella serovar sera were higher than the cut-off value, which indicated that established iELISA method could detect the Salmonella infection in ducks. The positive serotest results for the remaining common bacteria were all negative, indicating that the iELISA is specific for duck-derived Salmonella (Fig. 5).
Repeatability of the iELISA
Repeatability test results showed intragroup and intergroup coefficient of variation (CVs) less than 5.6 and 9.7%, respectively, indicating that the established iELISA method has good repeatability (Table 3).
Comparison of iELISA with dot-blot
A total of 100 serum samples were used to evaluate the potential of developed ELISA by comparison with Dot-blot. The developed iELISA showed 89% coincidence rate with the Dot-blot (Table 4).
Application of iELISA method to clinical samples
A total of 613 serum samples were collected from 3 different duck farms. All serum samples were monitored using the iELISA and commercial ELISA kit (Table 5). The results showed that 257 and 320 serum samples tested positive and negative, respectively. The analysis of discrepant data was obtained for 26 sera that was positive by iELISA but be negative by commercial ELISA kit. 10 sera were negative by iELISA but positive by commercial kit. Therefore, there were good agreement (94.1%) between iELISA and commercial kit.
Discussion
Duck industry is an important part of animal husbandry in China, and the quality of duck products is closely related to human health. Recent study has shown that 604 isolates of Salmonella from 3340 retail meat samples from 2009 to 2016, of which 241 strains were isolated from poultry (ducks and chickens), accounting for 40% of the total isolates [25]. These showed that Salmonella infection in poultry has been very serious. Compared with chickens, little attention has been paid to ducks Salmonellosis. Therefore, there is an urgent need to determine the prevalence of Salmonella in ducks, which help curb the spread of this pathogen in duck farms.
So far, more than 2600 identified serotypes display different pathogenicities and an extensive host range [26]. Many studies have reported that ducks can be infected with multiple serotypes of Salmonella [27, 28], therefore the established method needs to be capable of broadly detecting various serotypes. Since there is no immunization policy for duck salmonellosis in China, we use antibody testing to confirm whether ducks are infected with Salmonella. iELISA is a simple, high throughput and strong specificity method that can be used for rapid pathogen detection, and is very suitable for the current domestic large-scale duck farms. Some studies have used lysed bacterial cells or LPS as the detection antigen to establish an iELISA kit. Although this method can detect many serotypes of Salmonella, it is easy to lead to false positive detection results because of antigenic epitopes similar to those on other bacteria, such as E. coli. The method developed by lipopolysaccharide (LPS) limited specificity of the antigen-antibody interaction. To solve this problem, we screened a special PagN protein, it is an outer membrane protein that contribute to the virulence, and is widely distributed and well conserved among the different species and subspecies of Salmonella. There are both B cell and T cell antigen peptides in its four extracellular rings, which can induce a strong immune response. In recent years, it has often been studied as a candidate protein for subunit vaccines [21, 29, 30].
In this study, E. coli prokaryotic expression system was used to express the recombinant protein in vitro. Although the expression system do not modify the protein, is easy to form inclusion body protein. Compared with other protein expression systems, the genetic background of E. coli expression system is clear, the operation process is relatively simple, easy to purify, and the protein expression production is high. Therefore, the expression system can effectively reduce the cost of the kit [31].
In this study, the complete DNA sequence of PagN gene was obtained by PCR, and cloned into pET-32a(+) to construct successfully the recombinant prokaryotic expression plasmid pET-32a(+)-PagN. There is a 6 × His tag protein in the pET-32a (+) plasmid itself, and the tag is fused and expressed at both ends of the PagN protein [32]. So the target protein can be efficiently purified by nickel ion affinity chromatography. SDS-PAGE analysis showed that the target protein with high purity was obtained in this study, which effectively eliminated the interference of E. coli bacterial protein, and further improved the specificity of the iELISA method established. Because the protein was in the form of inclusion body, 8 M urea was used to denature the protein in the purification process, resulting in no biological activity of the protein. The dialysis method with appropriate concentration gradient is very efficient for protein refolding and has almost no effect on protein concentration [33]. We used this method to refold the protein and finally obtained the soluble active protein.
We initially established the iELISA assay for detecting Salmonella antibodies based on recombinant PagN protein, and determined the optimal reaction conditions. In the present study, we obtained a fixed cut-off of 0.268 for iELISA performance by detecting 38 Salmonella negative serum samples. The method has high specificity sensitivity when treated with S. enteritidis, S. typhimurium and S. Kottbus positive sera from ducks. At the same time, iEILSA method had no cross-reaction with antibodies against E. coli, Staphylococcus aureus and Streptococcus. Compared with the iELISA method established using PagC protein [34], the method we established is more accurate for E. coli positive serum, indicating that the method established in this study has better specificity. Although the sensitivity of this study was lower than previous studies, the minimum detection limit of the referenced positive sera was still 1:1600, which was also applicable to sera with lower antibody titers. The specificity of the test may be higher when the sensitivity is low, and the specific relationship between sensitivity and specificity in this study needs to be supported by more subsequent experimental data. Because of the strong subjectivity of agglutination test, and we found that sometimes there was no agglutination between different serotypes of Salmonella and positive sera. Therefore, we used dot blot as similar to the principle of iELISA to detect serum samples, and the coincidence rate was 89%. The difference between the two methods may be due to the stronger specificity of iELISA. The use of the iELISA and commercial ELISA kit to detect the clinical serum samples at the same time demonstrated an agreement of 94.1%, which suggest that the iELISA based on PagN recombinant protein method can perform detection of Salmonella infection.
Although the established iELISA method is high in efficiency and sensitivity, it still requires further optimization. For example, the OD readings obtained with non-Salmonella sera were close to those of the cut-off value, suggesting a certain level of cross-reactivity. Cross-reactions can be further evaluated by detecting more non-Salmonella positive sera and expressing specific PagN epitopes.
Conclusion
In conclusion, an iELISA-base method using recombinant multiepitope PagN protein as the coating antigen was established successfully. This method has high sensitivity and specificity, and with no serological cross-reaction with other pathogens. We believed that the newly built iELISA method could be a useful detecting tool for large-scale monitoring the epidemiology of Salmonella infection in ducks.
Materials and methods
Strains, serum samples and antibodies
35 ducks at 7 days of age were selected to confirm Salmonella-free ducks by bacterial isolation and identification [35]. After these ducks were raised for 7 days, 5 ducks were randomly selected to collect blood and obtain negative serum control. The remaining ducks were randomly divided into 6 groups (5 per group) according to different pathogenic bacteria, including Salmonella enteritis, Salmonella typhimurium, Salmonella Cottbus, E. coli, Staphylococcus aureus and Streptococcus, above bacteria culture were assigned the 0.2 mL of 6.0 × 106 CFU/mL by three intramuscular injections at 7-day intervals. At week 1 after the last immunization, blood samples from three ducks of each group were used to separate the serum, and then the antibody level of each serum was determined by tube agglutination test [36]. All ducks used for preparing negative serum to Salmonella and positive serum to other bacteria were raised in separate isolators, deprived freely of food and water.
The salmonella positive serum from ducks in present study is from Harbin Animal Husbandry and Veterinary Research Institute, and the serum was identified by agglutination test.
Predicting and screening linear B-cell epitopes
Based on the sequence of the PagN gene, we predicted and screened linear B-cell epitopes using DNA Star software and IEDB website (http://www.iedb.org/). The antigenic index, surface probability and hydrophilicity values are screened.
PagN gene amplification and recombinant plasmid construction
Primers were designed to amplify PagN gene using NCBI reference sequences of Salmonella enteritis (CP050712.1). Upstream primer PagN-F: 5′-CGGGATCCAAAGAAGGGATCTATATCACCGG-3′, downstream primer PagN-R: 5′-CCCAAGCTTAAAGGCGTAAGTAATGCCGAGC-3′, underlined is BamHI and HindIII restriction site, respectively. Using Salmonella enteritis (CVCC3377) genomic DNA as a template, the PagN fragment was amplified by PCR using PagN specific primers, and the gene fragment was purified using a gel extraction kit. After digestion with BamHI and HindIII, the products were purified and ligated to pET-32a(+) by T4 DNA ligase at 16 °C for 8 h,and then transformed into E.coil DH5α competent cells. A single colony on the LB solid medium (containing 100 μg/mL ampicillin) was selected and cultured overnight at 37 °C with shaking at 220 rpm. The positive clones were verified by restriction enzyme digestion and DNA sequencing, yielding pET-32a (+)-PagN.
Expression and purification of the recombinant PagN protein
The recombinant plasmid pET-32a (+)-PagN was transformed into E.coli BL21(DE3) expression strain. Positive clones were picked up and inoculated into 5 mL LB liquid medium with 100 μg/mL of ampicillin overnight at 37 °C. The overnight culture was inoculated into 200 mL LB liquid medium at a ratio of 1:100 and incubated at 37 °C and 220 rpm until the logarithmic growth phase. Expression of recombinant protein was induced by addition of IPTG. The bacterial pellet was collected by centrifugation, the cells were resuspended in phosphate buffered solution (PBS) with 1/10 culture volume and disrupted by sonication. The supernatant and pellet after sonication were collected and subjected to SDS-PAGE to analyze the expression form of the recombinant protein. The recombinant PagN protein was purified using Ni2+-NTA agarose gel affinity chromatography. The bound protein was eluted with elution buffer (20 mmol/L Tris-HCL, 200 mmol/L imidazole). Finally, the target protein was renatured in a gradient of 6 M, 4 M, 2 M urea buffer solution at 4 °C and then the purified protein concentration was measured by BCA protein assay kit (Beyotime, Shanghai, China).
Western blotting assays
The purified recombinant PagN protein was subjected to polyacrylamide gel electrophoresis and then electrotransferred onto a Nitrocellulose (NC) membrane. The NC membrane was blocked at 4 °C in blocking buffer with Tris-buffered saline (TBS) contained 3% BSA and 0.1% Tween 20 (TBST), and then washed 3 times for 5 min in TBST. The blocked membrane was incubated in duck anti-Salmonella serum (dilution 1/500, from Harbin Animal Husbandry and Veterinary Research Institute) at 4 °C for 2 h. The membrane was washed 3 times with TBST, and 1:4000 diluted HRP-goat anti-duck IgG (KPL, Gaithersburg, USA) was added and incubated for 2 h at 4 °C. After washed three times, enhanced chemiluminescent (ECL) developer solution was added to develop the color.
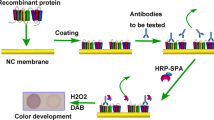
Establishment of the recombinant PagN protein-based iELISA
According to the classical iELISA method [17, 34], the optimal antigen coating concentration and serum dilution of the iELISA test reagents was investigated by checkerboard titration. Briefly, 100 μL of different concentrations (0.25, 0.5, 1.0, and 2.0 μg/mL) of PagN proteins were coated on a 96-well ELISA microplate overnight at 4 °C, 37 °C 1 h or 2 h, respectively. The ELISA plates were then washed three times for 5 min with washing buffer (PBS + 0.5% TWEEN-20). The wells were blocked with 200 μL blocking buffer (1% BSA, 2% BSA, 3% BSA, 3% skimmed milk, 4% skimmed milk and 5% skimmed milk) at 37 °C for 0.5 h, 1 h, 1.5 h, 2.5 h and 3 h, respectively. After three further washes, 100 μL serially diluted the positive and negative serum samples (2-fold dilutions, from 1:25 to 1:400) was added to each well and then incubated at 37 °C for 0.5 h,1 h, 1.5 h or 2 h. After three washes, the samples were incubated with 100 μL of goat anti-duck IgG-horseradish peroxidase conjugate (HRP) (KPL, Gaithersburg, USA) with two-fold serial dilutions (1:2000 to 1:8000) at 37 °C for 0.5 h, 1 h, 1.5 h or 2 h, washed again, and detected with 100 μL 3,3′,5,5′-Tetramethylbenzidine (TMB) color developer at room temperature for 5 min, 10 min,15 min or 20 min and away from light. The reaction was stopped by the addition of 50 μL 2 M H2SO4. All samples were set up in triplicate and measured with a microplate spectrophotometer (model 680, Bio-Rad) at 450 nm. The corresponding positive value (P) was approximately 1.0, the negative value (N) was less than 0.4, and the maximum difference in optical density (P/N) was not less than 2.1, which was considered to be the best reaction [37, 38].
Determination of the cut-off value
Under the best condition, the OD450 values of 38 negative serum of healthy ducks were determined by the iELISA method, and each serum sample was repeated three times. The mean and standard deviation of the OD450 values were calculated. The cut-off value was determined by titration as the mean OD450 (−x) value plus 3 the standard deviation (SD) of the antibody levels of 38 negative serum samples [34]. If the OD450 value of the test sample is higher than the cut-off value, the sample is regards as positive and vice versa.
Sensitivity analysis and specificity of iELISA test
To accurately assess the diagnostic sensitivity of the assay, the Salmonella positive serum was diluted from 1:50 to 1:12800. The highest dilution that produced an OD450 value> the cut-off value was considered as the detection limit of the iELISA assay (three replicates each test serum) [34]. In addition, to evaluate the diagnostic specificity, the iELISA method described above was used to simultaneously detect the OD450 values of positive serum antibodies against duck-derived Salmonella Enteritidis, Salmonella Typhimurium, Salmonella Cottbus, E. coli, Staphylococcus aureus and Streptococcus with three replicates each serum sample. The Salmonella- positive and -negative serum were set as controls.
Repeatability analysis
Under the optimal conditions established, three sero-positive serum samples and one sero- negative serum sample were detected by the iELISA. Four repeats were set for each sample, and the average value, standard deviation and coefficient of variation of each sample were calculated.
Comparison of iELISA with dot-blot
Dot-blot was used to validate the of the developed iELISA. iELISA and Dot-blot were used to detect 100 clinical sera at the same time, and the coincidence rate of them was compared. The steps for Dot-blot are as follows:
1 μg recombinant protein was fully adsorbed on NC membrane at 4 °C. The NC membrane were then washed 4 times for 5 min with TBST buffer. 5% skim milk was added to each well and incubated at 37 °C for 2 h. After 4 further washes, the NC membrane was placed in the positive serum diluted by 5% skimmed milk powder and incubated at 37 °C for 1 h. After 4 washes, the NC membrane was paced in the HRP goat anti-duck IgG by 5% skimmed milk powder and incubated at 37 °C for 1 h. Finally, after washing for 4 times, use enhanced diaminobenzidine (DAB) color developing solution to avoid light and develop 10 min.
Comparing with commercial ELISA test kits for detection of antibodies against Salmonella
To evaluate such iELISA kit using for detection of Salmonella in ducks, 613 clinical serum samples were tested by iELISA established in present study and a commercial ELISA kit (catalog number YJ660391;Shanghai Enzyme-linked Biotechnology Co., Ltd., China), which specifically designed to a double-antigen sandwich ELISA for detecting antibodies to Salmonella in serum by following the manufacturer’s instructions. A purified protein specific for Salmonella was used as a capture antigen and an HRP-conjugate for detecting the antibodies. Agreement between iELISA and the kit results was determined by counting the number of identical results and dividing it by the total number of samples.
Statistical analysis
Statistical analyses were performed using SPSS software (SPSS18.0). All values given in the text are the mean ± SD from the experiment.
Availability of data and materials
All data generated or analyzed during the current study are included in this published article.
Abbreviations
- iELISA:
-
Indirect enzyme-linked immunosorbent assays
- PCR:
-
Polymerase chain reaction
- qPCR:
-
Quantitative real-time
- PCR; LAMP:
-
Loop-mediated isothermal amplification
- ICTS:
-
Gold immunochromatographic test strip
- PBS:
-
Phosphate buffered solution
- TBS:
-
Tris-buffered saline
- TBST:
-
Tris-buffered saline Tween-20
- ECL:
-
Enhanced chemiluminescent
- HRP:
-
Horseradish peroxidase
- IPTG:
-
Isopropyl-b-D-thiogalactopyranoside
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel elctrophoresis
- NC:
-
Nitrocellulose
- TMB:
-
3,3′,5,5′-Tetramethylbenzidine
- LPS:
-
Lipopolysaccharide
- DAB:
-
Diaminobenzidine
References
Wei B, Cha SY, Kang M, Park IJ, Moon OK, Park CK, et al. Development and application of a multiplex PCR assay for rapid detection of 4 major bacterial pathogens in ducks. Poult Sci. 2013;92(5):1164–70.
Liljebjelke KA, Hofacr CL, Liu T, White DG, Ayers S, Young S, et al. Vertical and horizontal transmission of Salmonella within integrated broiler production system. Foodborne Pathog Dis. 2005;2(1):90–102.
Yang XJ, Huang JH, Wu QP, Zhang JM, Yang SY, Wang J, et al. Occurrence, serovars and antibiotic resistance of Salmonella spp. in retail ready-to-eat food products in some Chinese provinces. LWT-food. Sci Technol. 2022;154:112699.
Yang XJ, Huang JH, Zhang YX, Liu SR, Chen L, Xiao C, et al. Prevalence, abundance, serovars and antimicrobial resistance of Salmonella isolated from retail raw poultry meat in China. Sci Total Environ. 2020;713:136385.
Shao D, Shi Z, Wei J, Ma Z. A brief review of foodborne zoonoses in China. Epidemiol Infect. 2011;139(10):1497–504.
Kurtz JR, Goggins JA, McLachlan JB. Salmonella infection: interplay between the bacteria and host immune system. Immunol Lett. 2017;190:42–5.
Schneider T, Hahn-Löbmann S, Stephan A, Schulz S, Giritch A, Naumann M, et al. Plant-made Salmonella bacteriocins salmocins for control of Salmonella pathovars. Sci Rep-UK. 2018;8(1):1–10.
O'Bryan CA, Ricke SC, Marcy JA. Public health impact of Salmonella spp. on raw poultry: current concepts and future prospects in the United States. Food Control. 2022;132:108539.
Law JW, Ab Mutalib NS, Chan KG, Lee LH. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol. 2015;5:770.
Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galán JE, Ginocchio C, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992;6(4):271–9.
Huang CX, Mahboubat BY, Ding YF, Yang QL, Wang J, Zhou M, et al. Development of a rapid Salmonella detection method via phage-conjugated magnetic bead separation coupled with real-time PCR quantification. LWT-Food Sci Technol. 2021;142:111075.
Wan JJ, Zheng LP, Kong LY, Lu ZX, Tao Y, Feng ZY, et al. Development of a rapid detection method for real-time fluorescent quantitative PCR of Salmonella spp. and Salmonella Enteritidis in ready-to-eat fruits and vegetables. LWT-food. Sci Technol. 2021;149:111837.
Wen JP, Gou HC, Zhan ZQ, Gao Y, Chen ZQ, Bai J, et al. A rapid novel visualized loop-mediated isothermal amplification method for Salmonella detection targeting at fimW gene. Poult Sci. 2020;99(7):3637–42.
Shang YT, Ye QH, Cai SZ, Wu QP, Pang R, Yang SH, et al. Loop-mediated isothermal amplification (LAMP) for rapid detection of Salmonella in foods based on new molecular targets. LWT-Food Sci Technol. 2021;142:10999.
Wu YX, Wu MJ, Liu C, Tian YC, Fang SQ, Yang H, et al. Colloidal gold immunochromatographic test strips for broad-spectrum detection of Salmonella. Food Control. 2021;126:108052.
Tran LT, Nguyen TN. Production of recombinant antigens, polyclonal antibodies for development of an ELISA assay to detect Salmonella spp. in food. J Biotechnol. 2008;136:S757.
Mirhosseini SA, Fooladi AAI, Amani J, Sedighian H. Production of recombinant flagellin to develop ELISA-based detection of Salmonella Enteritidis. Braz J Microbiol. 2017;48:774–81.
Sannigrahi S, Arumugasamy SK, Mathiyarasu J, KS. Magnetosome-anti-Salmonella antibody complex based biosensor for the detection of Salmonella typhimurium. Mater Sci Eng C-Mater Biol Appl. 2020;114:111071.
Li A, Zuo P, Ye BC. An aptamer biosensor based dual signal amplification system for the detection of Salmonella typhimurium. Anal Biochem. 2021;615:114050.
Yang YJ, Wan CX, Xu HY, Aguilar ZP, Tan QL, Xu F, et al. Identification of an outer membrane protein of Salmonella enterica serovar typhimurium as a potential vaccine candidate for salmonellosis in mice. Microbes Infect. 2013;15(5):388–98.
Barilleau E, Védrine M, Koczerka M, Burlaud-Gaillard J, Kempf F, Grépinet O, et al. Investigation of the invasion mechanism mediated by the outer membrane protein PagN of Salmonella typhimurium. BMC Microbiol. 2021;21(1):153.
Lambert MA, Smith SG. The PagN protein of Salmonella enterica serovar typhimurium is an adhesin and invasin. BMC Microbiol. 2008;8(1):1–11.
Lambert MA, Smith SG. The PagN protein mediates invasion via interaction with proteoglycan. FEMS Microbiol Lett. 2009;297(2):209–16.
Bumann D. Identification of protective antigens for vaccination against systemic salmonellosis. Front Immunol. 2014;5:381.
Xu ZH, Wang M, Zhou CY, Gu GM, Liang J, Hou Z, et al. Prevalence and antimicrobial resistance of retail-meat-borne Salmonella in southern China during the years 2009-2016: the diversity of contamination and the resistance evolution of multidrug-resistant isolates. Int J Food Microbiol. 2020;333:108790.
LeLievre V, Besnard A, Schlusselhuber M, Desmasures N, Dalmasso M. Phages for biocontrol in foods: what opportunities for Salmonella sp. control along the dairy food chain? Food Microbiol. 2019;78:89–98.
Wang J, Li JX, Liu FL, Cheng YY, Su JL. Characterization of Salmonella enterica isolates from diseased poultry in northern China between 2014 and 2018. Pathogens. 2020;9(2):95.
Yang J, Ju ZJ, Yang Y, Zhao XN, Jiang ZY, Sun SH. Serotype, antimicrobial susceptibility and genotype profiles of Salmonella isolated from duck farms and a slaughterhouse in Shandong province. China BMC microbiol. 2019;19(1):1–12.
Goswami N, Hussain MI, Borah P. Molecular dynamics approach to probe the antigenicity of PagN -an outer membrane protein of Salmonella Typhi. J Biomol Struct Dyn. 2018;36(8):2131–46.
Das S, Chowdhury R, Ghosh S, Das S. A recombinant protein of Salmonella Typhi induces humoral and cell-mediated immune responses including memory responses. Vaccine. 2017;35(35):4523–31.
Lavallie ER, Mccoy JM. Gene fusion expression systems in Escherichia coli. Curr Opin Biotechnol. 1995;6(5):501–6.
Liu ZQ, Yang PC. Construction of pET-32a(+) vector for protein expression and purification. N Am J Med Sci. 2012;4(12):651–5.
Yamaguchi S, Yamamoto E, Mannen T, Nagamune T. Protein refolding using chemical refolding additives. J Biotechnol. 2013;8(1):7–31.
Ma Z, Yang XY, Fang YZ, Tong ZX, Lin HX, Fan HJ. Detection of Salmonella infection in chickens by an indirect enzyme-linked immunosorbent assay based on presence of PagC antibodies in sera. Foodborne Pathog Dis. 2018;15(2):109–13.
Mondal T, Khan MSR, Alam M, Purakayastha M, Das M, Siddique MP. Isolation, identification and characterization of Salmonella from duck. Bangl J Vet Med. 2008;6(1):7–12.
Sandhu T, Harry E. Serotypes of Pasteurella anatipestifer isolated from commercial White Pekin ducks in the United States. Avian Dis. 1981;25(2):497–502.
Naqvi MAUH, Naqvi SZ, Memon MA, Aimulajiang K, Haseeb M, Xu LX, et al. Combined use of indirect ELISA and western blotting with recombinant hepatocellular carcinoma-associated antigen 59 is a potential immunodiagnostic tool for the detection of prepatent Haemonchus contortus infection in goat. Animals. 2019;9(8):548.
Wu Y, Cheng A, Wang M, Zhang S, Zhu D, Jia AB, et al. Serologic detection of duck enteritis virus using an indirect ELISA based on recombinant UL55 protein. Avian Dis. 2011;55(4):626–32.
Acknowledgements
Not Applicable.
Funding
This work was supported by the Natural Science Foundation of Shandong Province of China [grant number ZR2020MC176], the Key Research and Development Program of Shandong Province, China [grant number 2022CXGC010606], and Shandong Modern Agricultural Technology & Industry System, China [grant number SDAIT-11-15].
Author information
Authors and Affiliations
Contributions
Conceived and wrote the pager: SH. Designed the experiment and modified the paper: YZ SJ. Performed the experiment and analyzed the data: SH SW XZ JG RZ. Collected samples: SW YW LG. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of Shandong Agricultural University (SDAUA-2017-029) and performed in accordance with the “Guidelines for Experimental Animals” of the Ministry of Science and Technology (Beijing, China), and ARRIVE guidelines (https://arriveguidelines.org/). We obtained informed consent from the owners of the duck flocks that were sampled, and the locations where we sampled are not privately owned or protected in any way.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hou, S., Wang, S., Zhao, X. et al. Establishment of indirect ELISA method for Salmonella antibody detection from ducks based on PagN protein. BMC Vet Res 18, 424 (2022). https://doi.org/10.1186/s12917-022-03519-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-022-03519-7