Abstract
Background
Glanders is a transmissible zoonotic disease caused by Burkholderia mallei that infects equids and humans. No glanders cases in equids were reported so far in Nepal.
Case presentation
Following suspected glanders in animals with clinical signs in different regions in Nepal, serum samples were tested by CFT, ELISA and Luminex® tests. Two horses and a mule tested positive for glanders by all tests, while two other equids only tested positive by ELISA and Luminex®. Analysis of swabs and pus samples by a PCR system targeting B. mallei confirmed the presence of the bacterium in the samples collected from the 3 equids that yielded positive results in all serological tests. Genotyping of the three PCR positive samples with a SNP-based method identified a genotype closely related to the B. mallei strains circulating in India.
Conclusion
Confirmation of glanders cases underscores the need of implementing a surveillance program in Nepal and a strict control of the animal movement across the borders.
Similar content being viewed by others
Background
Glanders is an infectious disease caused by Burkholderia mallei. This zoonotic bacterium primarily infects equids [1]. Several outbreaks of glanders in equids have been recently reported in South Asia, the Middle East, and South America (Brazil) [2]. Clinical and laboratory diagnosis of glanders is difficult since limited clinical signs are expressed in the early stage of infection. Symptoms of B. mallei infection include nasal discharge, pneumonia, and ulcerating nodular lesions on the skin. Discharges from the respiratory tract and skin are infectious. Transmission between animals is facilitated by close contact, inhalation, ingestion of contaminated materials (e.g., from infected feed and water troughs), or by inoculation (e.g., via a harness). Diagnostic methods of glanders include immunological tests such as complement fixation test (CFT) or ELISA and/or allergic reaction (malleinization), as well as direct tests such as bacteriological isolation and molecular tests. For B. mallei typing, the high-resolution melting PCR (HRM-PCR) technique targeting single nucleotide polymorphisms (SNPs) allows categorization of strains into three lineages (L1 to L3), as well as branches, sub-branches, and clusters with geographic specificities [3,4,5].
Case presentation
In November 2020, equids in Banke district of mid-western Nepal developed clinical signs and symptoms like high grade fever (up to 40-410C), labored breathing, dry cough, loss of appetite, lameness, thick mucopurulent yellowish nasal discharge, pus filled nodules on different parts of body, especially on thigh area. Later, in December 2020, similar clinical symptoms and signs were observed in mules in Dhading and Lalitpur districts of the Bagmati Province. The death of several equids was also reported in Nepalgunj, Lumbini province (Fig. 1). Available clinical surveillance data are reported in Table 1. Sticky yellowish pus discharge from ulcerated nodules and scabs was noticed on some of the infected equids (Fig. 2). Mules showed more severe symptoms than horses. All the infected animals were isolated and given symptomatic treatment. In most cases, symptoms relapsed after some time, became more severe and animals died of the disease. There is no policy to euthanize glanders infected animals in Nepal.

© Nations Online Project, https://www.nationsonline.org/oneworld/map/nepal-administrative-map.htm)
Map showing the geographical locations of the glanders-affected districts in Nepal, between November and December 2020 (map source:
For diagnosis and further investigations, sera and tissue samples were collected from three horses and two mules, each from different owners. One horse (L/157) came from the Lalitpur district, while the four other equids came from the Banke district (B/113, B/115, B/117 and B/120).
Sera from these five equids were analyzed with five different serological tests: (i) CFT [6], (ii) ID Screen® Glanders indirect ELISA (IdVet, France) [7], (iii) GLANDA ELISA (IdVet, France) based on two recombinant proteins [8], (iv) Luminex® bead-based assay targeting Hcp1 and GroEL proteins [9], and (v) ELISA based on a glycoengineered protein of Burkholderia recently validated for glanders diagnosis (Glyco ELISA) [10]. Two horses (L/157, B/120) and one mule (B/113) were positive by all tests and positive or undetermined results were obtained for all animals with ID Screen® Glanders indirect ELISA, GLANDA ELISA and Luminex® tests. The CFT and the ELISA test based on a glycoengineered protein identified fewer positive samples (Table 2).
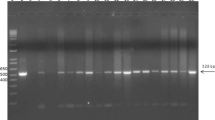
In parallel, nasal or pus swabs were collected from these five animals. Samples were submitted to DNA extraction and PCR amplifications as previously described [4]. Briefly, after DNA extraction with the High Pure PCR Template Preparation Kit (Roche, Meylan, France), DNA was amplified by real-time PCR using four different PCR systems: fliP (specific for B. mallei), orf11 (specific for B. pseudomallei), and aroA (specific for the B. pseudomallei complex). All samples with a quantification cycle (Cq) over 39 were considered as negatives. Both aroA and fliP PCR detected a positive signel for the three equids (L/157, B/113, B/120) that were positive by all serological tests (Table 3).
We further genotyped the B. mallei strain from these three PCR positive samples. After a pre-amplification step to increase the amount of template (using the Perfecta® pre-amplification kit (Quantabio) and the corresponding set of primers), DNA samples were analyzed by PCR-HRM [3]. The panel of 15 markers was used to classify the B. mallei strains into one of the three lineages (L1 to L3) and the branches, sub-branches, and groups to which they relate. All samples corresponded to the L2B2sB2branch, which includes B. mallei strains circulating in India and Pakistan. A recent study, using four new SNP markers, classified B. mallei strains from India and Pakistan into two small and large subgroups [5]. This new set of markers were investigated in the three positive Nepalese samples from Lalitpur (L/157) and Banke (B/113 and B/120). The results indicated that all samples clustered in the India_group 2 (large), which includes most of the Indian strains typed so far with this new set [5], all originating from the states of Uttar Pradesh and Haryana, in Northern India (Fig. 3). The origin of the India_group 1 (small) strains is not clearly defined at this time.
Discussion and conclusions
Recently Adhikari and colleagues [11] warned about the potential risks of glanders outbreaks in Nepal due to the re-emergence of the disease in neighboring Indian states [12,13,14], and the unrestricted movement of equids between the two countries. Most equids from India passed through the Nepalgunj quarantine office, the closest to India’s Uttar Pradesh region [11], where glanders cases are regularly reported [12,13,14]. Our epidemiological investigation indicates the equids were imported from Uttar Pradesh through uncontrolled routes, as there is about 1,770 kilometers of open border between India and Nepal. Further, there is seasonal migration of horses and mules from far western part of Nepal to western and central part and back to the India’s Uttar Pradesh region. Most of the equids in Nepal (an estimated number of 59,762; Ministry of Agriculture and Livestock Development (MOALD), 2019/2020) are used in the brick industry for transportation of bricks, goods and pulling carts. Equids are also popular in the tourism industry for transport of goods by trekkers. Horses are only vehicles for transport of goods and humans in high hill areas where mechanical vehicles cannot be used [15].
In May 2021, Nepal notified to OIE its first outbreak of glanders. Until now, no policy for prevention and control of this disease was implemented. The confirmation of glanders cases, detected in central and mid-western parts of Nepal and reported in this study, should prompt a national surveillance programme and enhanced border control measures. In general, the glanders situation in Asia is very poorly documented and strict measures are necessary to control this re-emergent disease.
Availability of data and materials
All data generated or analyzed during this study are included in this published article. The datasets generated during and/or analysed during the current study are available from the corresponding author.
Abbreviations
- CFT:
-
Complement fixation test
- DNA:
-
Desoxyribose Nucleic Acid
- iELISA:
-
Indirect ELISA
- ELISA:
-
Enzyme-Linked Immuno Assay
- HRM-PCR:
-
High Resolution Melting-PCR
- PCR:
-
Polymerase Chain Reaction
- SNP:
-
Single Nucleotide Polymorphism
References
Khan I, Wieler LH, Melzer F, Elschner MC, Muhammad G, Ali S, Sprague LD, Neubauer H, Saqib M. Glanders in animals: a review on epidemiology, clinical presentation, diagnosis and countermeasures. Transbound Emerg Dis. 2013;60:204–21.
World Organization for Animal Health. 2018. Terrestrial animal health code. Available at: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.05.11_GLANDERS.
Girault G, Wattiau P, Saqib M, Martin B, Vorimore F, Singha H, Engelsma M, Roest HJ, Spicic S, Grunow R, Vicari N, De Keersmaecker SCJ, Roosens NHC, Fabbi M, Tripathi BN, Zientara S, Madani N, Laroucau K. High-resolution melting PCR analysis for rapid genotyping of Burkholderia mallei. Infect Genet Evol. 2018;63:1–4.
LaroucauK AR, Vorimore F, Varghese K, Deshayes T, Bertin C, Delannoy S, SamiAM AB, M, El ShorbagyM, AlmutawaaKAW, SaadAlanezi SJ, Alazemi YSN, Guernier- CambertV, Wernery U. A genetic variant of Burkholderia mallei detected in Kuwait: Consequences for the PCR diagnosis of glanders. Transbound Emerg Dis. 2021;68(2):960–3.
Singha H, Vorimore F, Saini Sheetal, Deshayes T, Saqib M, Tripathi BN, Laroucau K. Molecular epidemiology of Burkholderia mallei isolates from India (2015–2016): New SNP markers for strain tracing. Infect Genet Evol. 2021;95:105059.
Laroucau K, Colaneri C, Jaÿ M, Corde Y, Drapeau A, Durand B, Zientara S, Beck C, Union E, laboratories involved in glandersserodiagnosis. Interlaboratory ring trial to evaluate CFT proficiency of European laboratories for diagnosis of glanders in equids. Vet Rec. 2016;178(25):632.
Elschner MC, Melzer F, Singha H, Muhammad S, Gardner I, Neubauer H. Validation of a commercial glanders ELISA as an alternative to the CFT in international trade of equidae. Front Vet Sci. 2021;8:628389.
Elschner M, Laroucau K, Singha H, El-Adawy H, Khan I, Saqib M, Melzer F, Gardner I, Neubauer H. Evaluation of the prescribed complement fixation test in comparison with western blot and five enzyme linked immune assays for serological diagnosis of glanders. PLoS One. 2019;14(4):e0214963.
Laroucau K, Saqib M, Martin B, Deshayes T, Bertin C, Wernery U, Joseph S, Singha H, Tripathi BN, Beck C. Development of a microsphere-based immunoassay for the serological detection of glanders in equids. Acta Topica. 2020;207:105463.
Wang G, Glaser L, Scott NE, Fathy Mohamed Y, Ingram R, Laroucau K, Valvano MA. A glycoengineered antigen exploiting a conserved protein O-glycosylation pathway in the Burkholderia genus for detection of glanders infections. Virulence. 2021;12(1):493–506.
Adhikari N, Acharya KP, Wilson RT. The potential for an outbreak of glanders in Nepal. Tropical Medicine and Health. 2019;47:57.
Malik P, Singha H, Khurana SK, Kumar R, Kumar S, Raut AA, Riyesh T, Vaid RK, Virmani N, Singh BK, Pathak SV, Parkale DD, Singh B, Pandey SB, Sharma TR, Chauhan BC, Awasthi V, Jain S, Singh RK. Emergence and re-emergence of glanders in India: a description of outbreaks from 2006 to 2011. Vet Ital. 2012;48(2):167–78.
Malik P, Singha H, Goyal SK, Khurana SK, Tripathi BN, Dutt A, Singh D, Sharma N, Jain S. Incidence of Burkholderia mallei infection among indigenous equines in India. Vet Rec Open. 2015;2(2):e000129.
Singha H, Shanmugasundaram K, Tripathi BN, Saini S, Khurana SK, Kanani A, Shah N, Mital A, Kanwar P, Bhatt L, Limaye V, Khasa V, Arora R, Gupta S, Sangha S, Sharma H, Agarwal SK, Tapase J, Parnam S, Dubey P, Baalasundaram SK, Mandal BN, Virmani N, Raj Gulati B, Malik P. Serological surveillance and clinical investigation of glanders among indigenous equines in India from 2015 to 2018. Transbound Emerg Dis. 2020. https://doi.org/10.1111/tbed.13475.
Bhatt BR, Kafle A, Shrestha S, and Kaphle K.2020. Horse Breeds of Nepal. International Journal of Zoology and Animal Biology. 10.23880/izab-16000216. https://www.moald.gov.np/publication/Agriculture%20Statistics
Acknowledgments
Not applicable
Funding
This project was supported by the Government of Nepal and by the European Commission’s Directorate-General for Health and Consumers.
Author information
Authors and Affiliations
Contributions
K.P., M.M., M.S., and P.K.R. contributed to the collection of samples and performed the clinical and preliminary diagnosis of these first cases of glanders in Nepal. D.T., W.G., V.M.A and K.L performed the complementary diagnosis investigations and molecular typing analysis. All authors contributed to the revising of the manuscript. The authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Samples from naturally infected animal were collected as part of routine veterinary investigation carried out by qualified veterinarians from the Central Veterinary Laboratory of the Government of Nepal.
Consent for publication
Not applicable
Competing interests
The authors have no financial or personal relationships with any individuals or organizations that could inappropriately influence or bias this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
P, K., M, M., S, M. et al. First glanders cases detected in Nepal underscore the need for surveillance and border controls. BMC Vet Res 18, 132 (2022). https://doi.org/10.1186/s12917-022-03233-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-022-03233-4