Abstract
Background
Staphylococcus aureus (S. aureus), especially methicillin-resistant Staphylococcus aureus (MRSA), is considered a common zoonotic pathogen, causing severe infections. The objective of this study was to investigate the antimicrobial susceptibility, resistance genes and molecular epidemiology among MRSA and methicillin-susceptible Staphylococcus aureus (MSSA) isolated from food animals in Sichuan Province, China.
Methods
This study was conducted on 236 S. aureus isolates. All isolates were subjected to antimicrobial susceptibility testing by using a standard microbroth dilution method. The Polymerase Chain Reaction (PCR) was performed to identify genes encoding the β-lactams resistance (blaZ, mecA), macrolides (ermA, ermB, ermC) and aminoglycosides (aacA-aphD). The molecular structures and genomic relatedness of MRSA isolates were determined by staphylococcal chromosome cassette mec (SCCmec) typing and pulsed-field gel electrophoresis (PFGE), respectively.
Results
Among 236 isolates, 24 (10.17 %) were recognized as MRSA. MRSA isolates showed different resistance rates to 11 antimicrobials ranging from 33.33 to 100 %, while for MSSA isolates the rates varied from 8.02 to 91.51 %. Multi-drug resistance phenotype was found in all MRSA isolates. The ermC gene encoding macrolides-lincosamides-streptogramin B was the most prevalent gene detected in 87.29 % of the S. aureus isolates, followed by ermB (83.05 %), blaZ (63.98 %), aacA-aphD (44.07 %), ermA (11.44 %) and mecA (11.02 %) genes. The prevalence of resistance genes in MRSA isolates was significantly higher than that of MSSA. Regarding the molecular morphology, SCCmec III (12/24, 50 %) was the most common SCCmec type. Furthermore, the PFGE typing showed that 24 MRSA were divided into 15 cluster groups (A to O), the major pulsotype J encompassed 25 % of MRSA isolates.
Conclusions
The S. aureus isolates from food animals in Sichuan province of China have severe antimicrobials resistance with various resistance genes, especially MRSA isolates. Additionally, the genetic pool of MRSA isolates is diverse and complex, and further investigation is necessary.
Similar content being viewed by others
Background
Staphylococcus aureus (S. aureus) is one of the important potential pathogens that can cause acute and chronic diseases, such as mastitis, endocarditis, sepsis, bacteremia, and toxic shock syndrome [1]. It has been reported that S. aureus can cause mastitis in dairy cows, dermatitis and sepsis in pigs, septic arthritis and subdermal abscesses in poultry [2]. Antibiotic therapy plays an important role in controlling infections caused by S. aureus. However, excessive use of antibiotics has resulted the development of resistant S. aureus strains, especially methicillin-resistant Staphylococcus aureus (MRSA), posing severe threat to humans and animals health [3]. Methicillin-susceptible Staphylococcus aureus (MSSA) generally evolves to MRSA through the acquisition and insertion of staphylococcal chromosome cassette mec elements (SCCmec), which can transfer horizontally and carry resistance gene mecA that codes penicillin-binding protein (PBP2a) with low affinity for β-lactam antibiotics [4]. The first occurrence of MRSA was reported in 1961, and the prevalence of MRSA increased year by year. The outbreaks of MRSA has been reported in many countries in the 1980s with high morbidity and mortality [5, 6]. In the USA, MRSA infection has been recognized in more than 320,000 cases and caused more than 10,000 deaths in 2017 [7]. Meanwhile, over 50 % of nosocomial S. aureus were associated with MRSA in most of the Asian countries [8]. In China, although the methicillin resistance rate had a trend of reduction since 2005, the prevalence of MRSA was still high [9, 10]. According to CHINET surveillance system in 2019, 31.4 % of all clinical isolates had been recognized as MRSA [10].
Apart from human studies, MRSA has also been known to exist in animals for a long time. MRSA colonies and infections have been reported in domestic livestock, companion animals and wildlife [11]. Prolonged misuse and abuse of antibiotics at farms largely contributed to the wide distribution of MRSA among food animals. It has been revealed that more than 40 % of pigs, 20 % of cattle, and 20–90 % of turkey farms have been affected by MRSA in Germany [12, 13], about 23–32 % of pig farmers were colonized with MRSA in swine farms in the Netherlands [14, 15]. In North America, the prevalence of MRSA had been found to be about 20 % [16]. Whereas in China, the prevalence of MRSA in dairy cow, pigs, and chicken ranches were 6.6 %, 49 %, and 2.1-3.5 %, respectively [17,18,19]. MRSA in food animals may cause not only animal diseases but also a zoonotic issue between animals and humans through direct contact, environmental contamination, and contaminated animal products [20, 21]. Overall, these findings suggested that the resistance and epidemiological studies of MRSA isolated from animals are necessary for both animal and human health.
Sichuan province is one of the largest producers of food animals, and areas for the production and use of animal drugs. However, there are scant studies regarding the prevalence of MRSA in food animals in Sichuan province. The purpose of the study was to evaluate resistance phenotypes and genotypes of MRSA and MSSA isolates among 236 S. aureus isolates from livestock and poultry in Sichuan province. Furthermore, the molecular types of MRSA isolates were analyzed by staphylococcal chromosome cassette mec (SCCmec) typing and pulsed-field gel electrophoresis (PFGE).
Results
Antimicrobial susceptibility testing
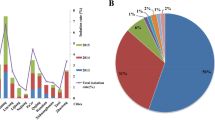
There were 236 S. aureus isolates isolated from sick chickens (n = 97), ducks (n = 124), swine (n = 11) and cows (n = 4). 24/236 (10.17 %) were MRSA and 212/236 (89.83 %) were MSSA. MRSA isolates showed different resistance rates to 11 antimicrobials ranging from 33.33 to 100 %. The serious resistance was not only to β-lactam antibiotics, but also to erythromycin and sulfafurazole (100 % resistance rate each). MRSA isolates were significantly resistant to all other antibiotics except for penicillin, tetracycline, and ciprofloxacin (P < 0.05 or P < 0.01) compared with MSSA isolates (Fig. 1). Furthermore, 100 % of MRSA isolates were multi-drug resistant whereas only 80.66 % of MSSA showed multi-drug resistance (Fig. 2).
Comparison of resistance of MRSA and MSSA isolates to antimicrobials. PEN, penicillin; AMP, ampicillin; OXA, oxacillin; GEN, gentamicin; KAN, kanamycin; AMK, amikacin; TET, tetracycline; ERY, erythromycin; AZM, azithromycin; CIP, ciprofloxacin; SIZ, sulfafurazole. * indicates significant difference (P<0.05), ** indicates extremely significant difference (P<0.01) of resistance rate between MRSA and MSSA
Detection of resistance genes in S. aureus isolates
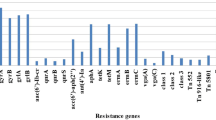
A total of 236 S. aureus isolates were tested for six antibiotic resistance genes including the β-lactamases blaZ gene (blaZ), methicillin resistance determinant (mecA), erythromycin ribosome methylase genes (ermA, ermB, ermC) and the bifunctional aminoglycoside N-acetyltransferase and aminoglycoside phosphotransferase (aacA-aphD) gene (Table 1). Among these genes, the main genotypes were ermC, ermB and blaZ (detection rate > 60 %). All MRSA isolates harboured mecA, ermB and ermC genes, while blaZ, ermA and aacA-aphD were detected in 87.50 %, 79.17% and 70.83 % of MRSA, respectively. Subsequent statistical analysis showed that prevalence of all resistance genes in MRSA isolates was significantly higher than that of MSSA (P < 0.05 or P < 0.01). As shown in Table 2, aacA-aphD-positive MRSA isolates showed resistance to aminoglycosides (gentamicin, kanamycin, amikacin) suggesting that the phenotype was in accordance with the genotype. In addition, all MRSA isolates harbored four or more resistance genes in this study. The most common multiple resistance gene combination profile was blaZ/mecA/ermA/ermB/ermC/aacA-aphD (54.17 %, 13/24) (Table 2)
Molecular typing of MRSA isolates
The genetic spectrum of the MRSA isolates was varied as revealed by the two typing approaches (Table 2). The characterization of the SCCmec cassettes revealed four different types in MRSA isolates: type I, III, IV, and V. SCCmec III was identified as the main SCCmec type, accounting for 50 % (12/24). Nevertheless, SCCmec I, IV, V and unidentified types were detected in 20.83 % (5/24), 8.33 % (2/24), 12.50 % (3/24) and 8.33 % (2/24), respectively. Non-typeable (NT) types were defined as isolates showing unexpected fragments. PFGE of SmaI-digested genomic identified the presence of 15 pulsotypes designated as A to O. PFGE pulsotype J was the largest one, encompassing 25 % (6/24) of isolates, followed by pulsotypes E, H, L and N, each containing two isolates. Each of the remaining pulsotypes (A, B, C, D, F, G, I, K, M, O) contained one isolate (Figs. 3 and 4). As shown in Table 2, the isolates belonging to pulsotype J were all identified as SCCmec III, suggesting the results of the two methods were consistent. But compared with SCCmec typing, PFGE distinguished the MRSA strains more specifically and detected more types.
The gels images of MRSA isolates by PFGE typing. a: 2, BMD18; 3, BMD24; 4, XCCow3; 5, YAD4; 6, YAD2; 7, GLD51; 8, GLD54; 9, YAC4; 10, BMD41; 11, BMD74; 12, BMD30; 13, BMD17; 14, YAD3; 1 and 15, Xba-digested DNA of Salmonella Braenderup H9812 used as DNA molecular size marker; b: 2, BMD42; 3, GLD59; 5, YAC1; 6, BMD68; 7, BMD23; 8, GLD83; 9, YAC2; 10, GLD93; 11, BMD9; 12, BMD20; 13, BMD69; 4, error (The forth lane was made by mistake.); 1 and 14, same as the 1 and 15 in a
Discussion
Inappropriate empirical use of antibiotics is a major cause of aggravation of drug resistance and poor curative effect. The emergence of MRSA in food animals associates with the presence of MRSA in human consumption foods. Further exploring the mechanisms of resistance and molecular epidemiology of S. aureus from animals is a key process in alleviating this crisis. In this study, antimicrobial susceptibility, resistance genes, and molecular epidemiology of the S. aureus isolated from food animals in Sichuan Province, China, were characterized.
In order to do a thorough screening of MRSA, we typed each isolate with both phenotype and genotype. The prevalence of MRSA in this study was 10.17 %, which is lower than the 14.2 % in Xinjiang [22], China, the 23.3 % in Brazil [23], and the 13 % in Denmark [24], whereas it is higher than that of the 6.8 % of a previous study in Sichuan [25], the 6.6 % in other provinces of China [17], the 9.2 % in Italy [26], and the 9.8 % in Germany [27]. The difference of prevalence may be due to different regions that were studied or the different species and physical status (healthy or sick) of the animals. Except for oxacillin, S. aureus isolates in this study showed high resistance to the antibiotics tested. The high resistance rates imply the misuse of antimicrobials in farms. However, oxacillin and amikacin, which were less used in veterinary medicine, still showed drug resistance, which indicates the potential of cross-resistance. Similar to the findings of Meng et al. [22] and Zayda et al. [28], MRSA isolates were found to be resistant to a broader spectrum of antimicrobials than MSSA isolates in the current study. Resistance rates of the MRSA isolates were significantly higher to gentamicin, kanamycin, amikacin, erythromycin, azithromycin and sulfafurazole compared with those of MSSA isolates, attributing to the existence of SCCmec in MRSA strains so that MRSA anchored more resistance genes than MSSA [29, 30]. However, there were no significant differences in the resistance rates of penicillin, tetracycline and ciprofloxacin between MRSA and MSSA (P > 0.05). The penicillin resistance rate was extremely high in both MSSA and MRSA isolates. The long-term use of antimicrobials is one of main driving force towards antimicrobial resistance. The reason behind the stable resistance rate maybe because penicillin, being a commonly used drug, was widely preferred for the treatment of Staphylococcal infections for a long time and established considerably stable resistance in S. aureus. Hence, the majority of S. aureus (either MRSA or MSSA) harbour the penicillinase encoded by blaZ that can hydrolyse penicillin. Multi-drug resistance (MDR) was defined as resistance to three or more families of antibiotics. Further comparison of MDR in MRSA and MSSA isolates, MDR was identified in all MRSA isolates and the proportion of multi-drug resistant strains in MRSA was almost three times higher than that of MSSA, which was in line with a previous study [31]. All these findings indicate the severity of antimicrobial resistance of MRSA isolates, which may be attributed to the increase in affinity of MRSA in acquiring mobile genetic elements (MGEs), such as transposons or conjugative plasmids carrying antimicrobial resistance genes [28, 30].
In our study, the majority of isolates possessed 1 to 6 resistance genes. The ermC gene was the most common one, which was detected in 87.29 % of S. aureus isolates. This finding is consistent with a previous report in China [32], but is different from the another study [33]. The ermB and blaZ genes were detected in 83.05 and 63.98 % of S. aureus isolates, respectively. These results indicate that the majority of S. aureus have the ability to acquire the three detected genes. In accord with previous studies [22, 32], a higher proportion of six genes encoding antibiotic-resistance were detected in MRSA isolates than in MSSA isolates. The prevalence of ermA and aacA-aphD in MRSA was significantly higher than that in MSSA (P < 0.01). Nearly 1 % (0.94 %) of isolates harboured the mecA gene but showed sensitivity to oxacillin and cefoxitin. This cryptic antibiotic-resistant S. aureus has been described in a Taiwanese study involving 91 S. aureus isolates with MIC 2.0 µg/ml, 57.1 % of which were mecA positive. The study also showed that 3.3 % of 180 S. aureus having 1 µg/ml MIC against oxacillin were mecA positive isolates [34]. Numerous genes may influence methicillin resistance phenotypes, for example the reason mecA-positive S. aureus showing sensitivity to oxacillin and cefoxitin may be because β-lactams regulatory genes affect the expression of resistance [35]. Strains with functional mec regulatory genes, such as mecI and mecR1 may produce little or no PBP2a, or the expressed protein may be inactivated, leading to a partially or completely suppressed expression of resistance [36, 37]. The ermB and ermC genes were detected in all MRSA isolates, and the gene blaZ was detected in all MRSA except three isolates, while they were only 81.13 %, 85.85% and 61.32 % in MSSA, respectively. Moreover, MRSA carried more abundant multidrug resistance genes compared with those of MSSA. Most of MSSA isolates carried two to four types, while all MRSA carried at least four resistance genes. The broader resistance gene spectrum of MRSA was consistent with its higher drug resistance compared with that of MSSA isolates, suggesting that the phenotype mostly reflects the resistance genotype.
In order to identify the genetic links of strains and to control MRSA infections effectively, it is important to master molecular characteristics. Several molecular typing methods for exploring molecular features of MRSA, such as PFGE, SCCmec, the staphylococcal protein A typing (spa), multilocus sequence typing (MLST), and so on are available [38,39,40]. Each of these techniques is used for specific purposes. SCCmec is used to recognize the structure and diversity of staphylococcal chromosome cassettes and can be classified as SCCmec type I to SCCmec XI [41]. MRSA strains may have different accessory genomes and carry different SCCmec elements [42]. Traditionally, healthcare-associated MRSA (HA-MRSA) carries SCCmec types I–III mainly, while community-associated MRSA (CA-MRSA) or livestock-associated MRSA (LA-MRSA) tend to harbor smaller SCCmec elements such as SCCmec type IV or type V [43, 44]. In the current study, MRSA isolates from the same areas and source showed the same SCCmec type, which may imply that the MRSA genotypes can be different in different regions. Most of the isolated MRSA were found to be SCCmec type III (12 isolates, 50 %), in accordance with the conclusion of a previous study that the predominant SCCmec type in Asia is type III [29]. However, the isolates in the current study were collected from animals, which may be a case of zoonosis. In addition, we found that the prevalence of combinatorial genotype blaZ/mecA/ermA/ermB/ermC/aacA-aphD was significantly higher in SCCmec type III, IV, and V, whereas this genotype was not detected in SCCmec type I. This finding was in line with the conclusion of a previous study that SCCmec type I carried fewer resistance genes [29]. PFGE, including enzyme restriction of bacterial DNA, separation of the restricted DNA bands and clonal assessment of bacteria, is a very sensitive approach for bacterial typing. Fifteen different clusters were obtained by PFGE typing in this study. Overall, no specific relationships had been identified between molecular features and origins. Genetic diversity was noted among animal species and regions, suggesting the complexity of genetic background of the MRSA isolates. All six MRSA isolates of cluster J were attributed to SCCmec type III. However, some MRSA isolates, such as YAD2, YAD3 and YAD4, from the same origins and with same resistance genes, were recognized as the same SCCmec type, while these were further discriminated into multiple types by PFGE. It has been revealed that PFGE is considered the “gold standard” for bacterial typing [45]. Results of our current study also indicated that the sensitivity of PFGE was higher than that of SCCmec typing method.
Conclusions
These findings revealed that the prevalence of antibiotic resistance of S. aureus from food animals is severe in Sichuan province, China, especially the MRSA isolates. MRSA isolates possess a broader spectrum of resistance genes than MSSA does. Additionally, the results of strain characterization suggest that the MRSA isolates from different origins and regions had genetic diversity and complex genetic background. The multiple resistance gene combination of blaZ/mecA/ermA/ermB/ermC/aacA-aphD was the most common combination profile in this study. The severity of drug resistance of these S. aureus isolates reflects the abuse of antibiotics in food animals. Therefore, it is of great significance to use antibiotics with caution and to strengthen the surveillance of MRSA at farms.
Methods
S. aureus isolates and identification of MRSA
A total of 236 S. aureus isolates were isolated from 13 locations of Sichuan Province, China between 2016 and 2019 (Fig. 5). All isolates were obtained from infected food animals including chickens (n = 97 isolates from 1246 samples), ducks (n = 124 isolates from 2155 samples), swine (n = 11 isolates from 148 samples) and cows (n = 4 isolates from 35 samples), sampling from articular exudates, livers, lungs and spleens of chickens and ducks with arthritis, milk of cows with mastitis, and skin swabs of pigs with skin infections and stored in an ice box after sampling for transportation. Samples were incubated at 37℃ in broth containing 1 % tryptone, 7.5 % sodium chloride, 1 % mannitol, and 0.25 % yeast extract for 22–24 h. S. aureus recognition was based on the growth status on Mannitol salt agar and CHROMagar™ Staph aureus medium, Gram-staining and standard biochemical tests. Only one isolate per animal sample was chosen for further analysis. The presence of MRSA isolates was confirmed by phenotypic identification methods screening for oxacillin and cefoxitin resistance [46], followed by polymerase chain reaction for detection of mecA [47]. The isolates were stored in -80 °C freezer until analysis.
Antimicrobial susceptibility testing of MRSA and MSSA
Antimicrobial MICs for MRSA and MSSA isolates were determined by broth microdilution and interpreted according to the CLSI guideline [46]. The antimicrobial agents included: penicillin G (PEN), ampicillin (AMP), oxacillin (OXA), gentamicin (GEN), kanamycin (KAN), amikacin (AMK), tetracycline (TET), erythromycin (ERY), azithromycin (AZM), ciprofloxacin (CIP) and sulfafurazole (SIZ). Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 25923 (BeNa Culture Collection, Beijing) were used as control strains. Multi-drug resistance (MDR) was defined as resistance to 3 or more families of antibiotics.
PCR amplification and sequencing of resistance genes of MRSA and MSSA
PCR was used to amplify the β-lactams (blaZ, mecA), macrolides (ermA, ermB, ermC) and aminoglycosides (aacA-aphD) antibiotic resistance genes. Six pairs of primers involved in the PCR reaction (Table 3), the mecA primers were cited from a previous study [47], the primers blaZ, ermA, ermB, ermC and aacA-aphD were designed by software Primer5. E.Z.N.A.™ Bacterial DNA Kit and Plasmid Mini Kit I (OMEGA) were used to extract bacterial DNA according to the manufacturer’s instruction. PCR amplification reactions were conducted in a total volume of 25µL with 1µL of the primer at concentration of 10µmol/L, 12.5µL of 2×Taq PCR MasterMix (TaKaRa, Dalian Co. Ltd), 1µL DNA template, and 9.5µL sterile deionized water. PCR amplification was carried out as follows: 5 min initial denaturation at 95℃, 30 cycles of denaturation at 95℃ for 30 s, annealing for 45 s (see annealing temperature for each gene in Table 3), extension at 72℃ for 45 s and final extension at 72℃ for 15 min. PCR products were analyzed by electrophoresis on 2 % agarose gel containing 0.5 µg/ml of etidium bromide in 0.5X TBE buffer, and the sequencing was determined by a commercial company (Qingke Biotechnology, Chengdu). The DNA sequences were analyzed by the BLAST program, available at the NCBI homepage (http://www.ncbi.nlm.nih.gov/BLAST/).
Molecular typing of MRSA isolates
SCCmec typing for the tested MRSA isolates was determined by the multiplex PCR method described by elsewhere [48]. All MRSA were analyzed by PFGE for their genetic relatedness. PFGE analysis of MRSA isolates tested in the study was as follows: the culture of strains, preparation of agarose gel, DNA digestion by SmaI and electrophoresis, all of which were practiced according to the protocol described by Bannerman et al. [49]. The PFGE banding patterns were interpreted with BioNumerics version 6.0 (Applied Math) by using UPGMA algorithm [50].
Statistical analysis
Statistical significance for the comparison of resistance rate was determined using X2- test by software SAS9.0. P < 0.05 was considered to be statistically significant.
Availability of data and materials
The DNA sequences were analyzed by the BLAST program, available at the NCBI homepage (http://www.ncbi.nlm.nih.gov/BLAST/).
Abbreviations
- S. aureus :
-
Staphylococcus aureus
- MRSA:
-
Methicillin-resistant Staphylococcus aureus
- MSSA:
-
Methicillin-susceptible Staphylococcus aureus
- WHO:
-
World Health Organization
- MDR:
-
Multi-drug resistance
- MGEs:
-
Mobile genetic elements
- PEN:
-
Penicillin G
- AMP:
-
Ampicillin
- OXA:
-
Oxacillin
- GEN:
-
Gentamicin
- KAN:
-
Kanamycin
- AMK:
-
Amikacin
- TET:
-
Tetracycline
- ERY:
-
Erythromycin
- AZM:
-
Azithromycin
- CIP:
-
Ciprofloxacin
- SIZ:
-
Sulfafurazole
- CLSI:
-
Clinical and Laboratory Standards Institute
- MIC:
-
Minimum inhibitory concentration
References
Wang B, Muir TW. Regulation of virulence in Staphylococcus aureus: Molecular mechanisms and remaining puzzles. Cell Chem Biol. 2016;23(2):214–24.
Haag AF, Fitzgerald JR, Penadés JR. Staphylococcus aureus in animals. Microbiol Spectr. 2019;7(3):10.1128/microbiolspec.GPP3-0060-2019.
Petinaki E, Spiliopoulou I. Methicillin-resistant Staphylococcus aureus among companion and food-chain animals: impact of human contacts. Clin Microbiol Infect. 2012;18(7):626–34.
Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44(6):1549–55.
Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31(4):e00020-18.
Mendem SK, Gangadhara TA, Shivannavar CT, Gaddad SM. Antibiotic resistance patterns of Staphylococcus aureus: A multi center study from India. Microb Pathog. 2016;98:167–70.
CDC. Antibiotic resistance threats in the United States, 2019. Atlanta: US Department of Health and Human Services, CDC. 2019.
Lim WW, Wu P, Bond HS, Wong JY, Ni K, Seto WH, et al. Determinants of methicillin-resistant Staphylococcus aureus (MRSA) prevalence in the Asia-Pacific region: A systematic review and meta-analysis. J Glob Antimicrob Resist. 2019;16:17–27.
Hu FP, Guo Y, Zhu D, Wang F, Jiang XF, Xu Y, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005–2014. Clin Microbiol Infect. 2016;22 Suppl 1:S9-14.
Hu F, Wang M, Zhu D, Wang F. CHINET efforts to control antibacterial resistance in China. J Glob Antimicrob Resist. 2020;21:76–77.
Cuny C, Friedrich AW, Kozytska S, Layer F, Nubel U, Ohlsen K, et al. Emergence of methicillin-resistant Staphylococcus aureus (MRSA) in different animal species. Int J Med Microbiol. 2010;300(2–3):109–17.
Idelevich EA, Lanckohr C, Horn D, Wieler LH, Becker K, Rock R. Multidrug-resistant bacteria in Germany: The impact of sources outside healthcare facilities. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2016;59(1):113–23.
Köck R, Ballhausen B, Bischoff M, Cuny C, Becker K. The impact of zoonotic MRSA colonization and infection in Germany. Berl Munch Tierarztl Wochenschr. 2014;127(9–10):384–98.
Lucia vRMM, Van KPH, Kluytmans JA. Increase in a dutch hospital of methicillin-resistant Staphylococcus aureus related to animal farming. Clin Infect Dis. 2008;46(2):261–3.
Voss A, Loeffen F, Bakker J, Klaassen CHW, Wulf M. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis. 2005;11(12):1965–6.
Khanna T, Friendship R, Dewey C, Weese JS. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet Microbiol. 2008;128(3–4):298–303.
Li T, Lu H, Wang X, Gao Q, Dai Y, Shang J, et al. Molecular characteristics of Staphylococcus aureus causing bovine mastitis between 2014 and 2015. Front Cell Infect Microbiol. 2017;7:127.
Sun C, Chen B, Hulth A, Schwarz S, Ji X, Nilsson LE, et al. Genomic analysis of Staphylococcus aureus along a pork production chain and in the community, Shandong Province, China. Int J Antimicrob Agents. 2019;54(1):8–15.
Wang X, Tao X, Xia X, Yang B, Xi M, Meng J, et al. Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail raw chicken in China. Food Control. 2013;29(1):103–106.
Aqib AI, Ijaz M, Anjum AA, Malik MAR, Mehmood K, Farooqi SH, et al. Antibiotic susceptibilities and prevalence of Methicillin resistant Staphylococcus aureus (MRSA) isolated from bovine milk in Pakistan. Acta Trop. 2017;176:168–172.
Erwin V, Jan K. Livestock-associated Staphylococcus aureus CC398: Animal reservoirs and human infections. Infect Genet Evol. 2014;21(1):523–30.
Dan M, Yehui W, Qingling M, Jun Q, Xingxing Z, Shuai M, et al. Antimicrobial resistance, virulence gene profile and molecular typing of Staphylococcus aureus isolates from dairy cows in Xinjiang Province, northwest China. J Glob Antimicrob Resist. 2019;16:98–104.
Guimaraes FF, Manzi MP, Joaquim SF, Richini-Pereira VB, Langoni H. Short communication: Outbreak of methicillin-resistant Staphylococcus aureus (MRSA)-associated mastitis in a closed dairy herd. J Dairy Sci. 2016;100(1):726–30.
Tang Y, Larsen J, Kjeldgaard J, Andersen PS, Skov R, Ingmer H. Methicillin-resistant and -susceptible Staphylococcus aureus from retail meat in Denmark. Int J Food Microbiol. 2017;249:72–76.
Yang X, Liu J, Huang Y, Meng J, Lei G, Jia Y, et al. Prevalence, molecular characterization, and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus from different origins in Sichuan Province, China, 2007–2015. Foodborne Pathog Dis. 2018;15(11):705–10.
Luini M, Cremonesi P, Magro G, Bianchini V, Minozzi G, Castiglioni B, et al. Methicillin-resistant Staphylococcus aureus (MRSA) is associated with low within-herd prevalence of intra-mammary infections in dairy cows: Genotyping of isolates. Vet Microbiol. 2015;178(3–4):270–4.
Monecke S, Ruppelt A, Wendlandt S, Schwarz S, Slickers P, Ehricht R, et al. Genotyping of Staphylococcus aureus isolates from diseased poultry. Vet Microbiol. 2013;162(2–4):806–12.
Zayda MG, Masuda Y, Hammad AM, Honjoh K, Elbagory AM, Miyamoto T. Molecular characterisation of methicillin-resistant (MRSA) and methicillin-susceptible (MSSA) Staphylococcus aureus isolated from bovine subclinical mastitis and Egyptian raw milk cheese. Int Dairy J. 2020;104:104646.
Liu J, Chen D, Peters BM, Li L, Li B, Xu Z, et al. Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathog. 2016;101:56–67.
Deurenberg RH, Vink C, Kalenic S, Friedrich AW, Bruggeman CA, Stobberingh EE. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2007;13(3):222–35.
Bernier-Lachance J, Arsenault J, Usongo V, Parent R, Archambault M. Prevalence and characteristics of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) isolated from chicken meat in the province of Quebec, Canada. PLoS ONE. 2020;15(1):e0227183.
Wang D, Wang Z, Yan Z, Wu J, Ali T, Li J, et al. Bovine mastitis Staphylococcus aureus: Antibiotic susceptibility profile, resistance genes and molecular typing of methicillin-resistant and methicillin-sensitive strains in China. Infect Genet Evol. 2015;31:9–16.
Qu Y, Zhao H, Nobrega DB, Cobo ER, Han B, Zhao Z, et al. Molecular epidemiology and distribution of antimicrobial resistance genes of Staphylococcus species isolated from Chinese dairy cows with clinical mastitis. J Dairy Sci. 2019;102(2):1571–83.
Chen FJ, Huang IW, Wang CH, Chen PC, Wang HY, Lai JF, et al. mecA-Positive Staphylococcus aureus with low-level oxacillin MIC in Taiwan. J Clin Microbiol. 2012;50(5):1679–83.
Chambers HF. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10(4):781–91.
Kuwahara-Arai K, Kondo N, Hori S, Tateda-Suzuki E, Hiramatsu K. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP 2’ production. Antimicrob Agents Chemother. 1996;40(12):2680–5.
Ryffel C, Kayser FH, Berger-Bachi B. Correlation between regulation of mecA transcription and expression of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 1992;36(1):25–31.
Holmes A, Moore LSP, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387(10014):176–87.
Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37(11):3556–63.
Enright MC, P. DN, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–15.
Haaber J, Penades JR, Ingmer H. Transfer of antibiotic resistance in Staphylococcus aureus. Trends Microbiol. 2017;25(11):893–905.
Florence C, Angeles AM, Wannes V, Katleen H, Freddy H, Patrick B. Transmission dynamics of methicillin-resistant Staphylococcus aureus in pigs. Front Microbiol. 2013;4:57.
Tsai HC, Tao CW, Hsu BM, Yang YY, Chen JS. Multidrug-resistance in methicillin-resistant Staphylococcus aureus (MRSA) isolated from a subtropical river contaminated by nearby livestock industries. Ecotoxicol Environ Saf. 2020;200:110724.
Monecke S, Slickers P, Gawlik D, Müller E, Reissig A, Ruppelt-Lorz A, et al. Variability of SCCmec elements in livestock-associated CC398 MRSA. Vet Microbiol. 2018;217:36–46.
Neoh HM, Tan XE, Sapri HF, Tan TL. Pulsed-field gel electrophoresis (PFGE): A review of the “gold standard” for bacteria typing and current alternatives. Infect Genet Evol. 2019;74:103935.
CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement M100 Wayne: Clinical and Laboratory Standards Institute. 2020.
Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, Boylevavra S, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA. 1998;279(8):593–8.
Havaei SA, Ghanbari F, Rastegari AA, Azimian A, Khademi F, Hosseini NS, et al. Molecular typing of hospital-acquired Staphylococcus aureus isolated from Isfahan, Iran. Int Sch Res Notices. 2014;2014:185272.
Bannerman TL, Hancock G, Tenover FC, Miller JM. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J Clin Microbiol. 1995;33(3):551–5.
Wang W, Liu F, Zulqarnain B, Zhang CS, Ke MA, Peng ZX, et al. Genotypic characterization of methicillin-resistant Staphylococcus aureus isolated from pigs and retail foods in China. Biomed Environ Sci. 2007;30(8):570–580.
Acknowledgements
I would like to give my sincere gratitude to Dr. Liren Zhang, Department of Veterinary Clinical Science, College of Veterinary Medicine, Oklahoma State University, who gave me great help in language improvement.
Statements
All methods were carried out in accordance with relevant guidelines and regulations such as Clinical and Laboratory Standards Institute.
Funding
This work was supported by the study on the epidemiology of antimicrobial resistance of Escherichia coli from fecal and environmental sources of captive giant pandas (grant number XNYB18-07).
Author information
Authors and Affiliations
Contributions
TG and GS designed the experiments, performed the experiments, analyzed the data, prepared figures and tables, authored drafts of the paper, and approved the final draft; JL guided and reviewed the design of this study and edited the manuscript; HF and HT contributed in sample collection; WZ, LZ and LY contributed to the isolation, identification, and antimicrobial susceptibility testing of the tested strains; QY contributed to the detection of resistance genes by PCR. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All samples collected from animals were collected under the written informed consent of farm owners and every farmer was provided with a document explaining the purpose and method of sample collection. All experimental protocols of this study were approved by the Ethics Committee of Sichuan Agricultural University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Gan, T., Shu, G., Fu, H. et al. Antimicrobial resistance and genotyping of Staphylococcus aureus obtained from food animals in Sichuan Province, China. BMC Vet Res 17, 177 (2021). https://doi.org/10.1186/s12917-021-02884-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-021-02884-z