Abstract
Background
A novel Brucella strain closely related to Brucella (B.) melitensis biovar (bv) 3 was found in Croatian cattle during testing within a brucellosis eradication programme.
Case presentation
Standardised serological, brucellin skin test, bacteriological and molecular diagnostic screening for Brucella infection led to positive detection in one dairy cattle herd. Three isolates from that herd were identified to species level using the Bruce ladder method. Initially, two strains were typed as B. melitensis and one as B. abortus, but multiplex PCR based on IS711 and the Suis ladder showed that all of them to belong to B. melitensis, and the combination of whole-genome and multi-locus sequencing as well as Multi-Locus Variable numbers of tandem repeats Analysis (MLVA) highlighted a strong proximity within the phylogenetic branch of B. melitensis strains previously isolated from Croatia, Albania, Kosovo and Bosnia and Herzegovina. Two isolates were determined to be B. melitensis bv. 3, while the third showed a unique phylogenetic profile, growth profile on dyes and bacteriophage typing results. This isolate contained the 609-bp omp31 sequence, but not the 723-bp omp31 sequence present in the two isolates of B. melitensis bv. 3.
Conclusions
Identification of a novel Brucella variant in this geographic region is predictable given the historic endemicity of brucellosis. The emergence of a new variant may reflect a combination of high prevalence among domestic ruminants and humans as well as weak eradication strategies. The zoonotic potential, reservoirs and transmission pathways of this and other Brucella variants should be explored.
Similar content being viewed by others
Background
Brucellosis in cattle, which can be caused by B.abortus, B. melitensis and B. suis [1], can significantly impact productivity on beef and dairy farms, and it poses zoonotic risk to humans, in whom infection can cause severe illness. The last reported B. abortus infections in cattle in Croatia occurred in 1964, while B. melitensis infections in cattle were reported in 2008 in herds kept with infected sheep [2] and in 2019 in herds kept with infected goats [3]. Since 2011, Croatia has conducted a brucellosis eradication programme in cattle according to European Directive 64/432/EC. All animals older than 12 months are tested annually using the Rose Bengal test (RBT), and positive animals are further tested using complement fixation (CFT), as well as indirect and competitive ELISAs. Depending on the epidemiological situation, seropositive animals are tested using a brucellin skin test, tissues and organs (head, mammary and genital lymph nodes, uterus, spleen, udder, fetal membranes) and stomach contents, spleen and lung from foetuses are collected at slaughterhouse for bacteriological testing.
Within this eradication programme, potential Brucella isolates are identified to genus and species levels using a combination of classical biotyping, multiplex PCR and molecular genotyping methods, which can include Multi-Locus Variable number of tandem repeats Analysis (MLVA), Multi-Locus Sequence Typing (MLST) [1, 4,5,6,7] and Whole-Genome Sequencing (WGS).
Here we describe the identification of a dairy cow herd with brucellosis within the framework of the Croatian eradication programme. The disease was attributed to infection with B. melitensis bv. 3, except in one cow infected with a B. melitensis variant difficult to identify using standard classical and molecular methods. The emergence of this novel strain points to ongoing Brucella evolution in the western Balkans area, which may be due to the appearance of new reservoirs or vectors forced strain mutation and poor efficacy of eradication measures. Future studies should explore new reservoirs and zoonotic significance for this and other potential new Brucella variants in the region.
Case presentation
During routine annual testing within the national eradication programme, a dairy cow herd (12 cows, 7 heifers and 4 calves) with brucellosis was identified in October 2018 on a farm in the Croatian village of Katinovac (45°14′30.6″N 15°55′31.8″E), close to the northern border with Bosnia and Herzegovina. At the time of testing, animals were healthy and showed no clinical signs of brucellosis. Other animal species were not present on farm. A total of 19 animals aged 1 year and older were tested using the RBT. A total of 10 animals were identified positive by RBT and indirect ELISA, and 7 of these animals were also positive by the CFT and competitive ELISA. In November 2018, 19 animals older than 1 year were tested on the brucellin skin test. Positive reaction on brucellin was found in 5 previously seropositive cows, which were sent to the slaughterhouse, where samples were collected and analysed bacteriologically. During the post mortem inspection visible lesions were not recorded.
Epidemiological investigation showed that the Croatian herd had been brucellosis-free since 2013, and no new animals had been introduced since 2008. Since 2016, the farm practised artificial insemination with no recent history of abortions. The farm owner denied contact with other sheep or cattle herds and indicated that the herd was kept on pastures bordering Bosnia and Herzegovina. Their water source was the river Glina, a natural borderline in this area.
Brucella strains were isolated from milk and various tissues from three cows that were found without any clinical symptoms. The strains were classically biotyped as described [4] based on CO2 requirement, H2S production, oxidase and urease activity, growth on dyes, lysis by phages and agglutination with monospecific sera (Tables 1 and 2). This biotyping was performed at the Croatian National Reference Laboratory (Zagreb) and European Reference Laboratory (Maisons-Alfort, France). All three isolates could grow without CO2, they produced H2S and expressed oxidase, and they hydrolysed urea. They did not grow on basic fuchsin medium, and they triggered agglutination of anti-A and anti-M sera. These findings are consistent with B. melitensis bv.3. However, isolate 7 was lysed by Tbilisi phages at 104 routine test dilution (RTD).
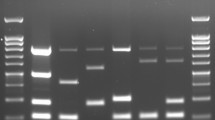
Species was determined using multiplex PCR based on the Bruce ladder [8] and the “AMOS” method [9], followed by the Suis ladder [10] and another PCR based on detection of the omp31 gene [11]. Isolates 6 and 11 gave results consistent with B.melitensis, but isolate 7 lacked omp31 gene sequences tested in the Bruce ladder, suggesting that it was B.abortus.
To confirm the identification of isolates 6 and 11 as well as to complete the identification of isolate 7, we performed MLVA based on 16 loci [5, 6] in the following order: panel 1, Bruce06 - Bruce08 - Bruce11 - Bruce12 - Bruce42 - Bruce43- Bruce45 - Bruce55; panel 2a, Bruce18 - Bruce19 - Bruce21; and panel 2b, Bruce04 - Bruce07 - Bruce09 - Bruce16 - Bruce30. B. melitensis 16M was used as the reference strain for comparison and verification of test quality. In addition, MLST was performed based on 9 loci [7]: gap - aroA - glk - dnaK - gyrB - trpE - cobQ - int-hyp (orf1)-omp25.
Moreover, isolates 6 and 7 were subjected to whole-genome shotgun sequencing using the Illumina NexteraXT system (protocol 150,319,425,031,942, revision C), which has been deposited in DDBJ/ENA/GenBank under accession numbers CVI_6 ChI CP058599/CVI_6 ChII CP058600 and CVI_7 ChI CP058597/CVI_7 ChII CP058598. A phylogenetic tree was generated using Bioumerics 7.6.3 (Applied Maths, BioMérieux). A set of B. melitensis genomes was retrieved from public databases (NCBI and PATRIC) and numbered in table (see Additional file 1). Sequencing reads were simulated for each genome using ART and all reads were mapped against a chimeric genome of B. melitensis 16M genome. Eight B. abortus genomes were used as outgroup. The SNPs obtained were then filtered (20X of absolute coverage, 10 bp inter-SNP distance, ambiguous and unreliable bases were removed, repeated elements removed) and a maximum parsimony tree was generated from these SNPs. The tree is represented with a logarithmic scale (see Additional files 2 and 3). This sequencing also revealed that isolate 7 contained a 609-bp omp31 sequence also present in isolate 6, but not the 723-bp omp31 sequence present in isolate 6. Based on classical and molecular methods, we assigned Brucella isolates 6 and 11 as B. melitensis bv. 3, while isolate 7 appeared to be a novel B. melitensis variant.
Discussion and conclusions
Human brucellosis, considered one of the most dangerous zoonoses, is most often caused by B. melitensis and less often by B. abortus or B. suis. The disease is endemic to the Mediterranean in general and the Balkan peninsula in particular [12,13,14,15,16]. Nevertheless, bovine brucellosis cases are sporadic and infrequent in Croatia, with the only recent reports limited to instances of transmission from sheep and goats on the same farms [2, 3]. The present case is the first recent report of brucellosis in Croatia that cannot definitively be attributed to contacts with other infected animal species.
This work highlights the need for continuing vigilance and research into potential Brucella reservoirs and spreading pathways. The disease in the present report may easily have come from Bosnia and Herzegovina, because herds on both sides of the border often share pastures, and illegal migrations are common which is documented throughout complete border line between countries [2, 3, 17]. Bosnia and Herzegovina has conducted a vaccination programme to control brucellosis in small ruminants since 2009, yet incidence of the disease remains high in animals and humans [18], and has even been increasing since 2012 [3]. This lack of efficacy is likely due largely to non-compliance with vaccination programmes [18], which can also foster the emergence of new Brucella strains [19].
We were unable to identify isolate 7 using classical microbiological methods [4] which are based on phenotype. This suggests that classical methods may not be well suited for characterising new B. melitensis strains in brucellosis-endemic regions. In fact, we were able to unambiguously identify the three isolates only by combining MLVA, MLST and whole-genome sequencing. These techniques showed our strains to be phylogenetically related to strains circulating in Croatia as well as Bosnia and Herzegovina [15, 17]. In particular, MLVA typing allowed us to assign a unique 16-digit code to the novel isolate 7, based on differences from the Bruce42 locus. Isolates 6 and 7 were assigned to the previously reported sequence type 8, related to B. melitensis strains circulating in Turkey, Kosovo and Macedonia. The two strains CVI_6 and CVI_7 clustered together in a subclade comprising 4 others strains (F9/05 from Turkey, BwIM_XXX_12 from unknown origin, F8/01–155 from Kosovo and BwIM_ALB_46 from Albania). Interestingly, this subclade contains two strains from the Balkan and one from Turkey. Moreover, in a recent paper [20], a strain from Serbia is clustered with the strain of Albania. The 2 strains isolated in this study seem to belong to a clade composed by strains that circulate in the Balkan area (see Additional files 2 and 3). Cross-contamination at the borders with animals can be a reason.
Our findings highlight the need for continuing, even enhanced, efforts to surveillance brucellosis in domestic animals and to research potential Brucella reservoirs and transmission pathways to ensure timely detection of zoonotic threats.
Availability of data and materials
All data generated during this study are included in this published article. The datasets of WGS genomes generated during the current study are available in the DDBJ/ENA/GenBank repository under accession numbers CP058599-CP058600 and CP058597-CP058598.
Abbreviations
- NCBI:
-
The National Center for Biotechnology Information advances science and health by providing access to biomedical and genomic information (https://www.ncbi.nlm.nih.gov/)
- PATRIC:
-
The Pathosystems Resource Integration Center is the all-bacterial Bioinformatics Resource Center (BRC) (http://www.patricbrc.org).
References
Manual of Diagnostic Tests and Vaccinesfor Terrestrial Animals 2019 (OIE): Brucellosis (Brucella abortus, B. melitensis and B. suis) (infection with B. abortus, B. melitensis and B. suis): https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.01.04_BRUCELLO-SIS.pdf. Accessed 31 Dec 2019.
Spicic S, Zdelar-Tuk M, Racic I, Duvnjak S, Cvetnic Z. Serological, bacteriological, and molecular diagnosis of brucellosis in domestic animals in Croatia. Croat Med J. 2010;51(4):320–6. https://doi.org/10.3325/cmj.2010.51.320.
World Animal Health Information System (WAHIS) - Version: 2: https://www.oie.int/wahis_2/public/wahid.php/countrymapinteractive. Accessed 31 Dec 2019.
Alton GG, Jones LM, Angus RD, Verger JM. Techniques for the brucellosis laboratory. Paris: Institut National de la Recherche Agronomique (INRA); 1988.
Le Flèche P, Jacques I, Grayon M, et al. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 2006;6(1):9. https://doi.org/10.1186/1471-2180-6-9.
Al Dahouk S, Flèche PL, Nöckler K, et al. Evaluation of Brucella MLVA typing for human brucellosis. J Microbiol Methods. 2007;69(1):137–45. https://doi.org/10.1016/j.mimet.2006.12.015.
Whatmore AM, Perrett LL, MacMillan AP. Characterization of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol. 2007;7(1):34. https://doi.org/10.1186/1471-2180-7-34.
López-Goñi I, García-Yoldi D, Marín CM, et al. Evaluation of a multiplex PCR assay (Bruce-ladder) for molecular typing of all Brucella species, including the vaccine strains. J Clin Microbiol. 2008;46(10):3484–7. https://doi.org/10.1128/JCM.00837-08.
Bricker BJ, Ewalt DR, MacMillan AP, Foster G, Brew S. Molecular characterization of Brucella strains isolated from marine mammals. J Clin Microbiol. 2000;38(3):1258–62. https://doi.org/10.1128/JCM.38.3.1258-1262.2000.
López-Goñi I, García-Yoldi D, Marín CM, de Miguel MJ, Barquero-Calvo E, Guzmán-Verri C, Albert D, Garin-Bastuji B. New Bruce-ladder multiplex PCR assay for the biovar typing of Brucella suis and the discrimination of Brucella suis and Brucella canis. Vet Microbiol. 2011;154(1–2):152–5. https://doi.org/10.1016/j.vetmic.2011.06.035.
Vizcaíno N, Verger JM, Grayon M, Zygmunt MS, Cloeckaert A. DNA polymorphism at the omp-31 locus of Brucella spp.: evidence for a large deletion in Brucella abortus, and other species-specific markers. Microbiology. 1997;143(9):2913–21. https://doi.org/10.1099/00221287-143-9-2913.
Krkic-Dautovic S, Mehanic S, Ferhatovic M, Cavaljuga S. Brucellosis epidemiological and clinical aspects (is brucellosis a major public health problem in Bosnia and Herzegovina?). Bosn J Basic Med Sci. 2006;6(2):11–5. https://doi.org/10.17305/bjbms.2006.3162.
Pappas G. The changing Brucella ecology: novel reservoirs, new threats. Int J Antimicrob Agents. 2010;36(S1):S8–S11. https://doi.org/10.1016/j.ijantimicag.2010.06.013.
Arapovic J, Spicic S, Ostojic M, et al. Epidemiological, clinical and molecular characterization of human brucellosis in Bosnia and Herzegovina - an ongoing brucellosis outbreak. Acta Med Acad. 2018;47(1):50–60. https://doi.org/10.5644/ama2006-124.214.
Duvnjak S, Racic I, Spicic S, Zdelar-Tuk M, Reil I, Cvetnic Z. Molecular epidemiology of Brucella melitensis strains causing outbreaks in Croatia and Bosnia and Herzegovina. Acta Vet Hung. 2018;66(2). https://doi.org/10.1556/004.2018.017.
Wareth G, et al. Brucellosis in the Mediterranean countries: history, prevalence, distribution, current situation and attempts at surveillance and control, Technical Series, vol. 12; 2019. ISBN 978–92–95115-00-2
Cvetnic Z, Zdelar-Tuk M, Duvnjak S, Racic I, Skrivanko M, Spicic S. Multiple locus variable number of tandem repeat analysis (MLVA) of isolates of Brucella melitensis isolated in the Republic of Croatia. Vet Arhiv. 2015;85:481–92.
Seric-Haracic S, Fejzic N, Saljic E, Hadzijunuzovic-Alagic D, Salman M. The scenario tree epidemiological model in estimation effects of B. melitensis rev. 1 vaccination on disease prevalence. Turk J Vet Anim Sci. 2018;42(5):416–4229. https://doi.org/10.3906/vet-1710-67.
Moreno E. Retrospective and prospective perspectives on zoonotic brucellosis. Front Microbiol. 2014;5:213. Published 2014 May 13. https://doi.org/10.3389/fmicb.2014.00213.
Georgi E, Walter MC, Pfalzgraf MT, Northoff BH, Holdt LM, Scholz HC, Zoeller L, Zange S, Antwerpen MH. Whole genome sequencing of Brucella melitensis isolated from 57 patients in Germany reveals high diversity in strains from Middle East. PLoS One. 2017;12(4):e0175425. https://doi.org/10.1371/journal.pone.0175425.
Acknowledgements
We thank Mrs. Silvija Drašković, Mrs. Marijana Novosel and Mr. Filip Radač from Croatian Veterinary Institute, NRL for Brucellosis for technical help.
Funding
This study was supported by Croatian Ministry of Agriculture and Croatian Veterinary Institute during collection, analysis, and interpretation of results within the framework of the Croatian eradication programme.
Author information
Authors and Affiliations
Contributions
MZT, SS, CP, IR and LF conducted serological and bacteriological analysis and interpreted data. SD, RSH, GG, PL performed the molecular examination of the strains including genotyping with result interpretation. VR, MR and SS conducted epidemiological analysis. All authors drafted and participated in writing of manuscript, read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All investigations were conducted as a part of regular disease-prevention-control activities and approved by Croatian Veterinary Directorate.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Table with strains metadata.
Additional file 2.
Textual description of phylogenetic tree.
Additional file 3.
Figure Phylogenetic Tree.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Spicic, S., Zdelar-Tuk, M., Ponsart, C. et al. New Brucella variant isolated from Croatian cattle. BMC Vet Res 17, 126 (2021). https://doi.org/10.1186/s12917-021-02833-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-021-02833-w