Abstract
Background
We aimed to investigate the prevalence, molecular epidemiology and prevalence factors for Extended Spectrum β-Lactamase-producing Enterobacteriaceae (ESBL-E) shedding by race horses. A cross-sectional study was performed involving fecal samples collected from 169 Thoroughbred horses that were housed at a large racing facility in Ontario, Canada. Samples were enriched, plated on selective plates, sub-cultured to obtain pure cultures and ESBL production was confirmed. Bacterial species were identified and antibiotic susceptibility profiles were assessed. E. coli sequence types (ST) and ESBL genes were determined using multilocus sequence type (MLST) and sequencing. Whole genome sequencing was performed to isolates harboring CTX-M-1 gene. Medical records were reviewed and associations were investigated.
Results
Adult horses (n = 169), originating from 16 different barns, were sampled. ESBL-E shedding rate was 12% (n = 21/169, 95% CI 8–18%); 22 ESBL-E isolates were molecularly studied (one horse had two isolates). The main species was E. coli (91%) and the major ESBL gene was CTX-M-1 (54.5%). Ten different E. coli STs were identified. Sixty-four percent of total isolates were defined as multi-drug resistant. ESBL-E shedding horses originated from 8/16 different barns; whereas 48% (10/21) of them originated from one specific barn. Overall, antibiotic treatment in the previous month was found as a prevalence factor for ESBL-E shedding (p = 0.016, prevalence OR = 27.72, 95% CI 1.845–416.555).
Conclusions
Our findings demonstrate the potential diverse reservoir of ESBL-E in Thoroughbred race horses. Multi-drug resistant bacteria should be further investigated to improve antibiotic treatment regimens and equine welfare.
Similar content being viewed by others
Background
Antibiotic resistance has been described as an emerging concern in a wide range of pathogens, such as extended spectrum β-lactamase-producing Enterobacteriaceae (ESBL-E) [1, 2]. ESBL are widespread enzymes, which confer resistance to extended spectrum cephalosporins and aztreonam, not to cephamycins or carbapenems, and are inhibited by β-lactamase inhibitors [3]. These genes are mostly mobile genetic element encoded, and may carry additional antimicrobial resistances, including aminoglycoside, sulfa-derivative, trimethoprim and quinolone resistance [4]. Therefore, treatment options are limited. In humans, infections with ESBL-E are associated with increased morbidity, mortality, length of hospital stay, delay of targeted appropriate treatment and higher costs [5].
Penicillins and cephalosporins are commonly prescribed in horses [6] and ESBL-E were reported as colonizing and infecting horses. In a longitudinal study in an equine clinic in the UK, increasing carriage rates were reported over a decade [7]. Reported infections in horses include wounds, respiratory, urinary tract and umbilical infections [8,9,10]. Race horses represent a unique equine community, due to high population density, shared training and living facilities, intense movement of horses, stress and medical treatments. All these factors create complex epidemiology for pathogen transfer [11]. Infectious diseases and bacterial pathogens specifically, have a major impact in the race horses industry leading to high rates of antibiotic use. Frequent inappropriate use of antimicrobials in race horses has been documented previously and implied that a greater scrutiny should be applied when determining whether antimicrobials are indicated in the treatment of horses with poor athletic performance [12]. Studies investigating antibiotic resistant bacteria in race horses are limited and have most often involved Enterococcus sp. [13], methicillin-resistant staphylococci [14] and E. coli [15]. A recent study detected 8.2% shedding rate of ESBL-producing E. coli in race horses [16]. However, data regarding colonization of race horses with different ESBL-E species, sequence types (ST) and factors associated with carriage are lacking.
In this study, we aimed to determine the ESBL-E shedding in Thoroughbred race horses, to identify the bacterial species, STs and ESBL genes, and to define the prevalence factors associated with ESBL-E shedding. We hypothesized that Thoroughbred race horses shed ESBL-E and that antibiotic treatment is a prevalence factor for shedding.
Results
Characterization of equine population
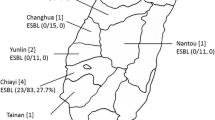
The study population included Thoroughbred horses originating from 16 different barns (Fig. 1), each barn trained by a different trainer. Between 2 and 35 horses were sampled per barn (mean = 11, median = 9), based on the owner’s or trainer’s permission, number of horses in the barn and number of horses from which feces were available. The classification of eight sampled horses to their specific barn was unavailable. Data regarding the identity of the barn’s veterinarian were available for 13 barns and included five different veterinarians. Medical records, other than signalment, were available for 56% of horses (n = 94/169). Age data were available for 104 horses, and the age ranged between two to 11 years, with a median of 3 years. Sex was recorded in 103 horses, including 46% females (n = 47/103), 42% geldings (n = 43/103) and 12% stallions (n = 13/103). Recent pathologies included orthopedic diseases, respiratory diseases, dermatological lesions, metabolic diseases, open wounds, colic, ophthalmic diseases and teeth abnormalities (Table 1).
Data regarding antibiotic treatment were obtained for 54% of horses (n = 91/169), of which 35% (n = 32/91) had been treated with an antibiotic at least once within the past year. Antibiotic treatments varied and included trimethoprim-sulpha, gentamicin, enrofloxacin, penicillin, ceftiofur, oxytetracycline and different combinations of these antibiotics. Data regarding omeprazole treatment was obtained for 54% of horses (n = 91/169), of which 35% (n = 32/91) were treated at least once within the past year. Data regarding anti-inflammatory treatment was obtained for 56% of horses (n = 95/169), of which 77% (n = 73/95) were treated at least once within the past year. Data regarding hospitalization and surgical procedures was obtained for 54% of horses (n = 92/169), of which 15% (n = 14/92) were hospitalized and/ or underwent a surgical procedure at least once within the past year (all parameters are divided to four time categories, as indicated in Table 1).
Prevalence of ESBL-E shedding
Twelve percent (n = 21/169) of the horses from 8/16 (50%) barns shed at least one ESBL-E strain. One horse shed two strains (Table 2). Overall, 22 ESBL-E were recovered. The majority of the ESBL-E isolates (n = 20/22, 91%) were E. coli, while the other ESBL-E isolates were identified as single isolates each of Enterobacter cloacae and Proteus hauseri.
Antibiotic susceptibility profiles, STs and blaESBL-genes
Antibiotic resistance rates of ESBL-E isolates were: 73% for trimethoprim-sulpha, 64% for tetracycline and gentamicin, 60% for chloramphenicol and 5% for nitrofurantoin. All isolates were resistant to ampicillin, piperacillin and cephalosporins (cefalexin, cefpodoxime, cefovecin and ceftifour), as well as susceptible to amikacin, ofloxacin, marbofloxacin and imipenem (Table 2). Sixty-eight percent of isolates (n = 15/22) were identified as multi-drug resistant (MDR).
Ninty-five percent of E. coli (n = 19/20) belonged to ten different STs and one E. coli isolate possessed a new combination of alleles suggesting a new E. coli ST. Ten STs included (Table 2): ST10 (a horse from barn 5, two horses from barn 6 and a horse from barn 8), ST1403 (three horses from barn 6), ST1730 (one horse from barn 8 and one horse from barn 12), ST1250 (one horse from barn 1, one horse from barn 5 and one horse from barn 6), ST86 (two horses from barn 6), ST1462 (one horse from barn 8), ST4527 (one horse from barn 5), ST7870 (one horse from barn 14), ST2008 (one horse from barn 6) and ST10787 (one horse from barn 16). Neither of the isolates belonged to the worldwide ESBL-producing E. coli ST131 lineage. The major ESBL gene found was blaCTX-M-1 (n = 12/22, 54.5%, Table 2).
Factors associated with ESBL-E shedding
In a mixed effects logistic regression, in which the barn was considered the random effect, shedding of ESBL-E by an individual animal was found to be significantly associated with antibiotic treatment within a month prior to sampling (p = 0.006, Table 1). Due to the small sample size, we chose to include the three most clinically relevant variables in the multivariable model [17]. Therefore, the model included the following parameters: antibiotic treatment within a month prior to sampling, surgery within a month prior to sampling (including two categories: castration and arthroscopy) and pathologies during three to six months prior to sampling (including two categories: respiratory and orthopedic lesions) (p ≤ 0.2, Table 3). The final model included 71 horses. The only significant prevalence factor in the model was antibiotic treatment within a month prior to sampling (p = 0.016, prevalence OR = 27.72, 95% CI 1.845–416.555, Table 3).
Discussion
Extended-spectrum β-lactamase-producing Enterobacteriaceae shedding by Thoroughbred race horses was investigated in this study, bacterial strains were characterized and a prevalence factor was determined.
Twenty-one out of 169 sampled horses (12%, 95% CI 8–18), originating from 16 different barns, shed ESBL-E. The prevalence of shedding horses varied between barns (zero to 33%), although there was no significant difference between barns, due to differences in the number of sampled horses. The difference in prevalence rates may be explained by the diversity of sampled facilities, representing different working environments (trainers, cleaners and veterinarians), different antibiotic use practices and transmission between barns. The overall prevalence we found in this study is higher compared to the rate of shedding ESBL-producing E. coli in an earlier study of UK equine farms [1] and similar to shedding rate in race horses with MDR E. coli reported from Korea [13]. In order to better understand whether ESBL-E colonization rate is significantly higher in race horses compared to farm horses, a cross sectional study should compare these two equine cohorts within the same country.
Drug resistance genes in general, and especially ESBL genes may originate from the environment, including manure, air and other animals. In recent studies, CTX-M-15 was identified as an emerging enzyme in human and in veterinary medicine in general and specifically in horses [7], but was not identified in this study. Drug resistance genetic variation may be also influenced by different facilities and interfaces [18]. The fact that a common gene, CTX-M-1, was detected in most horses (57%, n = 12/21) may also indicate on a common environmental source.
A high ESBL-E prevalence rate in Thoroughbred race horses may be a consequence of high stress and intensive interface, including a high number of horses housed and trained in the same facility and a frequent medication usage, especially antibiotics. A recent study on healthy farm horses revealed that more than five people taking care of horses daily, as well as the presence of an horse that has been treated medically on the last three months in the farm, were associated with ESBL/ Ampicillin C beta lactamase (AmpC) carriage [19]. These factors are relevant to the intensive care of race horses as well. A high prevalence rate in race horses has broader implications considering human health, animal health and welfare, as well as an ecological impact. Resistant bacterial dissemination may influence farm personnel, as owning or a contact with a horse was associated with ESBL-E carriage in humans [20]. Infections with these MDR bacteria limit treatment options; thereby affect both human and horse health and welfare. In this study, ESBL-E were investigated and detected in manure, but in a recent study it has also been detected in air samples and nostril swabs in farm horses [18]. A possible environmental ESBL-E contamination, originated from intensive equine facilities, may have an important ecological impact. Therefore, the importance of ESBL-E prevalence in these facilities is crucial.
The major ESBL-E species was E. coli (91%), followed by Enterobacter cloacae and Proteus hauseri (one isolate each). E. coli isolates belonged to multiple sequence types, which varied between and within the same facility. According to a previous study, intra- and inter-horse genetic diversity exists among E. coli strains and horizontal transfer and/ or selection of resistance genes probably occur within the equine gut microbiome [21]. Therefore, in our study, we suggest that the existence of diverse sequence types occurs due to horizontal transfer of β-lactamase genes and plasmids rather than clonal spread.
Among the ESBL-producing E. coli isolates recovered, we identified two E. coli strains, which are known to also infect people; ST10 and ST86. E. coli ST10 was identified before as a very common ST, isolated from human urinary tract infection cases and food-animal reservoirs in Canada [22], Europe [23] and the United States [24]. E. coli ST86 was also reported as a human pathogen, causing diarrhea and bacteremia [25, 26]. Both ST10 and ST68 were also isolated from horses in equine clinics in Europe, as well as ST1250 and ST2008 [9, 27, 28]. ST1730 has been also isolated before from equine clinical samples [28]. Overall, we identified some colonizing strains and some resistant zoonotic pathogens, which may pose a potential transmission risk to equine handlers. Five E. coli STs (ST10, ST1403, ST250, ST1730 and ST86) occurred in different barns during the study period suggesting inter-barn transmission. In barns 5, 6 and 8, multiple horses shed ESBL-producing E. coli, each colonized with a different ST (three to five different STs).
Moreover, the most prevalent ESBL gene we identified was CTX-M-1. In the last decade, the CTX-M gene family has become dominant and involved in many antibiotic resistant human infections in complicated community patients, usually with underlying disease, recent antibiotic usage or healthcare contact [29]. In equine studies, blaCTX-M-1 group was also reported as the most prevalent gene group associated with ESBL-production [9]. Transmission of plasmid carrying blaCTX-M-1 between commensal E. coli in pigs and farm workers was demonstrated before [30], a finding which reinforces the potential zoonotic risk. A further study should investigate the link between human-equine contact and bacterial transfer, as well as the duration of carriage following antibiotic treatment, in order to establish appropriate guidelines.
In light of our findings and the lack of data regarding ESBL-E in race horses, there is a great importance in identifying factors associated with ESBL-E shedding. We determined that individual antibiotic therapy is associated with ESBL-E shedding (p = 0.016), as described before both in dogs [31] and humans [32]. We did not identify any specific antibiotic agent which was significantly associated with shedding, probably due to multiplicity of agents in use. Therefore, a horse treated with antibiotics may constitute a risk for resistant bacterial transfer to its environment for an undetermined period of time. In addition, we revealed a variety of antimicrobials used in Thoroughbred race horses, including third generation cephalosporins and quinolones. A previous study revealed inappropriate use of antibiotics in race horses [12], which may explain the variety of antibiotics in use. We also identified that most of the isolates were MDR (68%, n = 15/22). In this study, we did not investigate clinical samples, or the connection between shedding and infection, which was demonstrated in human medicine before [33]. A further study should focus on clinical samples and inquire the connection between antibiotic treatment, ESBL-E shedding and infection in race horses, which has a financial significance.
Limitations of this study include convenience sampling and a relatively small sample size, which affect the ability to generalize these results. In this study, we were limited in sample size due to owners and trainers permission. In future studies, larger number of horses in more barns, should be selected randomly, which could result in higher statistical power, not only based on their availability on the sampling dates, but this must be determined in advance. Furthermore, it would be better to sample horses in more than one race-track and preferably in more than one country.
Another limitation is that this was a cross-sectional study and therefore descriptive and not longitudinal or cohort. Cross-sectional study are limited and cannot determine which variable is the cause and which the effect. For example, the finding that shedding of ESBL-E by an individual animal was found to be significantly associated with antibiotic treatment within a month prior to sampling. However, there is no guarantee that the ESBL-E bacteria were not present prior to antibiotic use, or even that clinical signs associated with ESBL-E did not lead to antibiotic use. Had this been a cohort study, the horses would have been verified to have been ESBL-E-free at the beginning of the study, which could have then led to the determination of risk factors influencing bacterial infection.
The use of retrospective medical data collection was another limitation of this study. This resulted in limited availability of medical records, which were only available for little over half of the horses. This further limit our ability to draw generalized conclusions and reveal associations since it lowered the sample size of the available information. A prospective study design in the future will enable to collect data for all horses and will enable to draw better conclusions.
Conclusion
The results from this study substantiate the occurrence of ESBL-E in Thoroughbred race horses. Our data confirms that Thoroughbred race horses may shed MDR ESBL-E, some of which may pose a zoonotic hazard. Recommendations for decreasing antimicrobial resistance among horses consist on implementation of antibiotic stewardship principle, including veterinary involvement, culture and sensitivity results, proper doses and overall decreased reliance on antimicrobials [34]. Further studies and active surveillance should focus on understanding ESBL-E epidemiology and transmission routes.
Methods
Equine study population, study design and sampling methods
This study was performed in a large racing facility in Ontario, Canada. The facility houses over 2000 horses from a large number of different trainers in 39 barns. Many horses move off-site regularly to race at other facilities or for other purposes.
Sample size was calculated (WinPepi, version 11.62) based on the assumption that the estimated prevalence of ESBL-production among potential pathogens in race horses is 10% [15], with power of 80%, and acceptable difference of 5%, resulting in minimal sample size of 139 horses. A convenience sample of horses was evaluated. All horses at the facility on the day of sampling were eligible for inclusion. Fresh fecal samples were collected during two sampling days in October 2017. Samples were collected from freshly passed feces in stalls, and only if it could be attributed to a specific horse. The number and selection of sampled animals at each barn was determined by owner’s or trainer’s permission. Overall, 169 fecal samples from 169 horses were collected. The study was performed in compliance with institutional guidelines for research on animals and an informed owner consent was obtained. Since we did not actually use horses an ethics approval was not required.
Equine demographic and medical data
Equine demographics and medical records were reviewed for the following information: signalment (age, sex and breed), veterinarian, barn/ trainer, pathologies, antibiotic treatment, omeprazole treatment, anti-inflammatory and anti-parasitic treatment, food additives, insertion of a nasogastric tube, endoscopy, hospitalization and surgical procedures. These parameters were recorded in four categories, depending on the time of treatment prior to sampling, (i) ≤ one month (ii) two to three months (iii) three to six months and (iv) six to twelve months.
Extended-spectrum β-lactamase-producing Enterobacteriaceae isolation and species identification
Fecal samples were collected in sterile containers and inoculated directly into an enrichment broth (BD Difco Luria Bertoni infusion enrichment broth, Becton Dickinson and Company, Sparks, USA) to increase sensitivity of ESBL-E detection [35]. After incubation at 37 °C (18-24 h), enriched samples were plated onto selective plates (Chromagar ESBL plates, Biomerieux Canada, St Laurent, Canada) and incubated at 37 °C for 24 h. When more than one colony was present, multiple colonies were isolated and tested only if they appeared different. Pure isolates after sub-culturing were stored at − 80 °C stocks for further analysis.
All isolates were subjected to analysis by Matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (MALDI-TOF, Microflex LT., Bruker Daltonics Ltd) for species identification and to vitek-2 analysis (BioMérieux, Inc., Marcy-l’Etoile, France) for antibiotic susceptibility testing using GN65 Vitek 2 card. Resistance rates reported were considered as complete resistance (excluding intermediate resistance). ESBL production was confirmed by clavulanic acid combination disk diffusion using cefotaxime and ceftazidime discs with and without clavulanic acid (BD BBL, Becton Dickinson and Company, Sparks, USA). Results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [36]. MDR bacteria were defined as such due to their in vitro resistance to three or more classes of antimicrobial agents [37].
Molecular characterization of ESBL-E
Isolates were examined for presence of the blaCTX-M ESBL groups using a multiplex polymerase chain reaction (PCR) from ESBL-E DNA lysates [37]. PCR was performed in a 10 μl mixture consisting of 2 μl GoTaq® Green Master Mix (Promega, Madison, USA), 0.3 μl DNase/RNase-free water (Promega, Madison, USA), 0.3 μl of each primer blaCTX-M-1, 1 μl of each primer blaCTX-M-2, 0.5 μl of each primer blaCTX-M-9 and blaCTX-M-25 (10 μM), 0.1 μl MgCl2 (50 mM) and 1 μl DNA template. PCR was carried under the following conditions: an initial 2 min denaturation at 94 °C, followed by 29 cycles of 30 s at 95 °C, 15 s at 61.7 °C and 30 s at 72 °C, with a final elongation at 72 °C for 5 min.
Isolates that were found to be blaCTX-M PCR negative were further examined for the presence of blaOXA-1, blaOXA-2, blaOXA-10 [38], blaTEM and blaSHV groups [39]. PCR for blaOXA genes was performed in different reactions (not multiplex) of 10 μl mixture consisting of 5 μl GoTaq® Green Master Mix, 3.6 μl DNase/RNase-free water, 1 μl of each primer and μl DNA template. PCR was carried under the following conditions: an initial 2 min denaturation at 94 °C, followed by 29 cycles of 30 s at 95 °C, 30 s at 51 °C/ 56 °C/ 55 °C for OXA-1/ OXA-2/ OXA-10 respectively and 60 s at 72 °C, with a final elongation at 72 °C for 5 min.
PCR for blaTEM and SHV genes was performed in two reactions of 50 μl mixture consisting of 25 μl GoTaq® Green Master Mix, 20 μl DNase/RNase-free water, 2 μl of each primer (Table S1) and 1 μl DNA template. PCR was carried under the following conditions: an initial 2 min denaturation at 95 °C, followed by 39 cycles of 95 °C for 1 min, 15 s at 52 °C and 50 s at 72 °C, with a final elongation at 72 °C for 5 min. PCR products were purified using a PCR purification kit, sequences were analyzed and compared with NCBI database to identify the ESBL gene allele.
All ESBL-producing E. coli Isolates were genotyped using an Enterobacterial repetitive intergenic consensus (ERIC) PCR amplification [40]. PCR was performed a 15 μl mixture consisting of 0.15 μl HIFI enzyme, 3 μl HIFI buffer (PCRBIOSYSTEMS, London, UK), 9.35 μl DNase/RNase-free water, 1.5 μl of ERIC2 primer and 1 μl DNA template. PCR was carried under the following conditions: an initial 30 s denaturation at 98 °C, followed by 34 cycles of 98 °C for 10 s, 30 s at 52 °C and 5 min seconds at 72 °C, with a final elongation at 72 °C for 10 min. Results were analyzed (GelJ, Java application) and strains showing a distinct ERIC PCR pattern were genotyped by multi locus sequence type (MLST) analysis as previously described using the Achtman scheme (IDGenomics, Seattle, USA) [41]. ST of two E. coli isolates (W65.1 and W112.1), for which ST were not identified by PCR- based MLST scheme, were determined in-silico using whole genome sequencing (WGS) data.
Whole genome sequencing
Twelve isolates, which were recognized as blaCTX-M-1 group-producers, were subjected for WGS, in order to determine in-silico the ST and the blaCTX-M-1 allele. The isolates were inoculated into LB agar with ampicillin (100 μg/m) at 37 °C for overnight incubation. Typical colonies from the fresh overnight culture were re-suspended in saline to about 1.5 OD600 nm. Four hundred microliter from each strain suspension was used for DNA extraction using MagNA Pure Compact (Roche) according to the manufacturer instructions. The DNA purity was determined using NaNodrop 2000 spectrophotometer and the amount of dsDNA was measured using DeNovix dsDNA High Sensitivity Kit (Cat# KIT-DSDNA-HIGH-2) and DeNovix QFX Fluorometer. DNA libraries were prepared using a Nextera XT DNA Library Preparation Kit following the manufacturer’s instructions. Sequencing was performed on an Illumina MiSeq platform using a 250-bp paired-end read v2 kit. Bioinformatics analyses were performed using the PATRIC v3.5.36 platform, including verification of the MLST profiles and CTX-M-1 genes [42]. The identification of CTX-M-1 gene alleles was confirmed by ResFinder-3.2 (https://cge.cbs.dtu.dk/services/ResFinder/).
Statistical analysis
Descriptive statistics were performed for all variables and are given as percentages, mean ± standard deviation or medians (range) (IBM Statistics SPSS, version 24.0).
To account for repeated measures within barns, logistic regression mixed-effect, with random effect at the barn-level, was used to explore the associations between the medical and demographic data and ESBL-E shedding.
Four variables (sex, veterinarian, pathologies and surgery) had more than two levels for comparison. The following variables were coded as yes/no (binary variables): antibiotic treatment, omeprazole usage, anti-inflammatory drugs usage, food additives, treatment with naso-gastric tube, endoscopy and hospitalization. These variables were evaluated at four different time frames (Table 1). Antibiotic treatment was coded as yes/no, due to variety of drugs and drug combination used (resulting in 10 different categories). The independent variables are stated in Table 1, whereas the dependent variable was ESBL-E shedding. A value of P ≤ 0.05 was considered significant. Multivariable analysis was conducted using selected variables with P ≤ 0.2 (StataCorp. 2017. Stata Statistical Software: Release 15. College Station, TX: StataCorp LLC).
Availability of data
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the Bio-Project accession number PRJNA612199.
Availability of data and materials
WGS data is available in the Genbank (Bio-Project accession number PRJNA612199), and the datasets used and analyzed during the current study are available from the corresponding authors on reasonable request.
Abbreviations
- AmpC:
-
Ampicillin C beta lactamase
- CHL:
-
Chloramphenicol
- CI:
-
Confidence interval
- CLSI:
-
Clinical and Laboratory Standards Institute
- CTX-M:
-
Cefotaxime hydrolyzing capabilities
- ENR:
-
Enrofloxacin
- ERIC:
-
Enterobacterial repetitive intergenic consensus
- ESBL:
-
Extended spectrum β lactamase
- ESBL-E:
-
Extended spectrum β lactamase-producing Enterobacteriaceae
- GENT:
-
Gentamicin
- MALDI-TOF:
-
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometer
- MAR:
-
Marbofloxacin
- MDR:
-
Multi-drug resistance
- MLST:
-
Multi locus sequence type
- NIT:
-
Nitrofurantoin
- OFL:
-
Ofloxacin
- OXA:
-
Oxacillin hydrolyzing capabilities
- SHV:
-
Sulfhydryl variable (enzyme’s active site)
- SPSS:
-
Statistical package for the social sciences
- ST:
-
Sequence type
- TEM:
-
Temoneira
- TET:
-
Tetracycline
- TMS:
-
Trimethoprim sulpha
- TOB:
-
Tobramycin
References
Maddox TW, Clegg PD, Diggle PJ, Wedley AL, Dawson S, Pinchbeck GL, et al. Cross-sectional study of antimicrobial-resistant bacteria in horses. Part 1: prevalence of antimicrobial-resistant Escherichia coli and methicillin-resistant Staphylococcus aureus. Equine Vet J. 2012;44:289–96.
Maddox TW, Pinchbeck GL, Clegg PD, Wedley AL, Dawson S, Williams NJ. Cross-sectional study of antimicrobial-resistant bacteria in horses. Part 2: risk factors for faecal carriage of antimicrobial-resistant Escherichia coli in horses. Equine Vet J. 2012;44:297–303.
Lee JH, Bae IK, Lee SH. New definitions of extended-spectrum β-lactamase conferring worldwide emerging antibiotic resistance. Med Res Rev. 2012;32:216–32.
Liu X, Thungrat K, Boothe DM. Occurrence of OXA-48 Carbapenemase and other β-lactamase genes in ESBL-producing multidrug resistant Escherichia coli from dogs and cats in the United States, 2009–2013. Front Microbiol. 2016;7:1057.
Schwaber MJ, Navon-Venezia S, Kaye KS, Ben-Ami R, Schwartz D, Carmeli Y. Clinical and economic impact of bacteremia with extended- spectrum-beta-lactamase-producing Enterobacteriaceae. Antimicrob Agents Chemother. 2006;50:1257–62.
Rubin JE, Pitout JDD. Extended-spectrum β-lactamase, carbapenemase and AmpC producing Enterobacteriaceae in companion animals. Vet Microbiol. 2014;170:10–8.
Isgren CM, Edwards T, Pinchbeck GL, Winward E, Adams ER, Norton P, et al. Emergence of carriage of CTX-M-15 in faecal Escherichia coli in horses at an equine hospital in the UK; increasing prevalence over a decade (2008–2017). BMC Vet Res. 2019;15:268.
Dierikx CM, van Duijkeren E, Schoormans AHW, van Essen-Zandbergen A, Veldman K, Kant A, et al. Occurrence and characteristics of extended-spectrum-β-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J Antimicrob Chemother. 2012;67:1368–74.
Walther B, Klein K-S, Barton A-K, Semmler T, Huber C, Wolf SA, et al. Extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Acinetobacter baumannii among horses entering a veterinary teaching hospital: The contemporary “Trojan Horse”. PLoS One. 2018:13.
Shnaiderman-Torban A, Paitan Y, Arielly H, Kondratyeva K, Tirosh-Levy S, Abells-Sutton G, et al. Extended-Spectrum β-lactamase-producing Enterobacteriaceae in hospitalized neonatal foals: prevalence, Risk Factors for Shedding and Association with Infection. Animals. 2019;9:600.
de la Rua-Domenech R, Reid SWJ, González-Zariquiey AE, Wood JLN, Gettinby G. Modelling the spread of a viral infection in equine populations managed in thoroughbred racehorse training yards. Prev Vet Med. 2000;47:61–77.
Weese JS, Sabino C. Scrutiny of antimicrobial use in racing horses with allergic small airway inflammatory disease. Can Vet J. 2005;46:438–9.
Kim DH, Chung YS, Park YK, Yang S-J, Lim SK, Park YH, et al. Antimicrobial resistance and virulence profiles of Enterococcus spp. isolated from horses in Korea. Comp Immunol Microbiol Infect Dis. 2016;48:6–13.
Mallardo K, Nizza S, Fiorito F, Pagnini U, De Martino L. A comparative evaluation of methicillin-resistant staphylococci isolated from harness racing-horses, breeding mares and riding-horses in Italy. Asian Pac J Trop Biomed. 2013;3:169–73.
Chung YS, Song JW, Kim DH, Shin S, Park YK, Yang SJ, et al. Isolation and characterization of antimicrobial-resistant Escherichia coli from national horse racetracks and private horse-riding courses in Korea. J Vet Sci. 2016;17:199–206.
Sukmawinata E, Sato W, Mitoma S, Kanda T, Kusano K, Kambayashi Y, et al. Extended-spectrum β-lactamase-producing Escherichia coli isolated from healthy thoroughbred racehorses in Japan. J Equine Sci. 2019;30:47–53.
Katz MH. Multivariable analysis: a primer for readers of medical research. Ann Intern Med. 2003;138:644–50.
Wolny-Koładka K, Lenart-Boroń A. Antimicrobial resistance and the presence of extended-spectrum beta-lactamase genes in Escherichia coli isolated from the environment of horse riding centers. Environ Sci Pollut Res. 2018;25:21789–800.
de Lagarde M, Larrieu C, Praud K, Schouler C, Doublet B, Sallé G, et al. Prevalence, risk factors, and characterization of multidrug resistant and extended spectrum β-lactamase/AmpC β-lactamase producing Escherichia coli in healthy horses in France in 2015. J Vet Intern Med. 2019;33(2):902–11.
Huijbers PMC, de KM, Graat EAM, van HAHAM, van SMG, de JMCM, et al. Prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in humans living in municipalities with high and low broiler density. Clin Microbiol Infect. 2013;19:E256–9.
Sadikalay S, Reynaud Y, Guyomard-Rabenirina S, Falord M, Ducat C, Fabre L, et al. High genetic diversity of extended-spectrum β-lactamases producing Escherichia coli in feces of horses. Vet Microbiol. 2018;219:117–22.
Manges AR, Harel J, Masson L, Edens TJ, Portt A, Reid-Smith RJ, et al. Multilocus sequence typing and virulence gene profiles associated with Escherichia coli from human and animal sources. Foodborne Pathog Dis. 2015;12:302–10.
Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect. 2011;17:873–80.
Yamaji R, Friedman CR, Rubin J, Suh J, Thys E, McDermott P, et al. A Population-Based Surveillance Study of Shared Genotypes of Escherichia coli Isolates from Retail Meat and Suspected Cases of Urinary Tract Infections. mSphere. 2018;3:e00179–18.
Guiral E, Pons MJ, Vubil D, Marí-Almirall M, Sigaúque B, Soto SM, et al. Epidemiology and molecular characterization of multidrug-resistant Escherichia coli isolates harboring blaCTX-M group 1 extended-spectrum β-lactamases causing bacteremia and urinary tract infection in Manhiça, Mozambique. Infect Drug Resist. 2018;11:927–36.
Kim JS, Kim J, Kim S-J, Jeon S-E, Oh KH, Cho S-H, et al. Characterization of CTX-M-type extended-spectrum beta-lactamase-producing diarrheagenic Escherichia coli isolates in the Republic of Korea during 2008-2011. J Microbiol Biotechnol. 2014;24:421–6.
Apostolakos I, Franz E, Hoek V, AHA M, Florijn A, Veenman C, et al. occurrence and molecular characteristics of ESBL/AmpC-producing Escherichia coli in faecal samples from horses in an equine clinic. J Antimicrob Chemother. 2017;72:1915–21.
Lupo A, Haenni M, Saras E, Gradin J, Madec J-Y, Börjesson S. Is blaCTX-M-1 riding the same plasmid among horses in Sweden and France? Microb Drug Resist. 2018;24:1580–6.
Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, et al. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother. 2007;59:165–74.
Moodley A, Guardabassi L. Transmission of IncN plasmids carrying blaCTX-M-1 between commensal Escherichia coli in pigs and farm workers. Antimicrob Agents Chemother. 2009;53:1709–11.
Belas A, Salazar AS, da GLT, Couto N, Pomba C. Risk factors for faecal colonisation with Escherichia coli producing extended-spectrum and plasmid-mediated AmpC β-lactamases in dogs. Vet Rec. 2014;175:202.
Ben-Ami R, Rodríguez-Baño J, Arslan H, Pitout JDD, Quentin C, Calbo ES, et al. A multinational survey of risk factors for infection with extended-Spectrum β-lactamase-producing Enterobacteriaceae in nonhospitalized patients. Clin Infect Dis. 2009;49:682–90.
Bert F, Larroque B, Paugam-Burtz C, Dondero F, Durand F, Marcon E, et al. Pretransplant fecal carriage of extended-Spectrum β-lactamase–producing Enterobacteriaceae and infection after liver transplant, France. Emerg Infect Dis. 2012;18:908–16.
Raidal SL. Antimicrobial stewardship in equine practice. Aust Vet J. 2019;97:238–42.
Jazmati N, Hein R, Hamprecht A. Use of an enrichment broth improves detection of extended-Spectrum-Beta-lactamase-producing Enterobacteriaceae in clinical stool samples. J Clin Microbiol. 2016;54:467–70.
CLSI, editor. Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI supplement M100. Wayne: Clinical and Laboratory Standards Institute; 2017.
Falagas ME, Karageorgopoulos DE. Pandrug resistance (PDR), extensive drug resistance (XDR), and multidrug resistance (MDR) among gram-negative bacilli: need for international harmonization in terminology. Clin Infect Dis. 2008;46:1121–2.
Lin S-P, Liu M-F, Lin C-F, Shi Z-Y. Phenotypic detection and polymerase chain reaction screening of extended-spectrum β-lactamases produced by Pseudomonas aeruginosa isolates. J Microbiol Immunol Infect. 2012;45:200–7.
Tofteland S, Haldorsen B, Dahl KH, Simonsen GS, Steinbakk M, Walsh TR, et al. Effects of phenotype and genotype on methods for detection of extended-spectrum-beta-lactamase-producing clinical isolates of Escherichia coli and Klebsiella pneumoniae in Norway. J Clin Microbiol. 2007;45:199–205.
Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–31.
Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–51.
Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother. 2006;57:154–5.
Acknowledgements
We acknowledge horses’ owners, barns’ employees and veterinarians for their collaboration in executing this study, as well as Dr. Gila Sutton for her assistance in the statistical analysis.
Funding
The authors would also like to thank the Hebrew University of Jerusalem for funding the travel expenses of AST to Ontario Veterinary College, University of Guelph, Canada.
Author information
Authors and Affiliations
Contributions
AST contributed to data collection, interpretation of data, microbiological and molecular analysis, as well as manuscript production. SNV contributed to the microbiological and molecular analysis, manuscript production and supervision. YP contributed to microbiological analysis. HA contributed to samples and data collection. WAA contributed to statistical analysis. DB contributed to study design and data collection. EH contributed to graphic design. SJW contributed to study design, statistical analysis, manuscript production and supervision. IN & GZV performed the WGS and contributed to the bioinformatics analysis. AS contributed to study design, manuscript production and supervision. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was performed in compliance with institutional guidelines for research on animals and an informed owner verbal consent was obtained. Permission was granted from the Woodbine Entertainment Group to carry out the collection of fecal samples from various shed rows at Woodbine Racetrack. Woodbine Entertainment Group is the operator of Woodbine Racetrack and provided us with a map of all barns on the grounds to facilitate the collection.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
PCR primers and reactions conditions
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Shnaiderman-Torban, A., Navon-Venezia, S., Paitan, Y. et al. Extended spectrum β lactamase-producing Enterobacteriaceae shedding by race horses in Ontario, Canada. BMC Vet Res 16, 479 (2020). https://doi.org/10.1186/s12917-020-02701-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-020-02701-z