Abstract
Background
Salmonella is an important zoonotic pathogen, and chickens are one of its main hosts. Every year, Salmonella infections pose a serious threat to the poultry industry in developing countries, especially China. In this study, a total of 84 Salmonella isolates recovered from sick and healthy-looking chickens in central China were characterized by serotyping, MLST-based strain typing, presence of potential virulence factors, and antimicrobial resistance profiles.
Result
Data showed that the main serotypes of Salmonella isolates in central China were Salmonella enterica serovar Gallinarum biovar Pullorum, Salmonella enterica serovar Gallinarum biovar Gallinarum, Salmonella enterica serovar Enteritidis and Salmonella enterica serovar Typhimurium, and among them, S. Pullorum was the dominant type in both sick and healthy-looking chickens, accounting for 43.9 and 46.5%, respectively, while S. Enteritidis was only found in healthy-looking chickens. All isolates exhibited higher resistance rates to ampicillin (97.6%), tetracycline (58.3%) and colistin (51.2%), and among these isolates, 49.5% were resistant to more than three drugs in different combinations. S. Enteritidis was the most severe multidrug-resistant serotype, which showed higher resistance rates to colistin, meropenem and ciprofloxacin. Multilocus sequence typing (MLST) revealed that S. Gallinarum and S. Enteritidis isolates were clustered in clade 1, which belonged to two and one STs, respectively. All S. Typhimurium isolates were clustered in clade 3, and belonged to three STs. However, S. Pullorum were distributed in three clades, which belonged to 7 STs. Twenty-seven virulence-associated genes were detected, and expected cdtB, which was absent in all the isolates, the other 26 genes were conserved in the closely related Salmonella serogroup D (S. Enteritidis, S. Pullorum, and S. Gallinarum).
Conclusion
Salmonella serogroup D was the major subgroup, and S. Pullorum was the most common type in sick and healthy-looking chickens in central China. Drug resistance assays showed serious multiple antimicrobial resistances, and S. Enteritidis was the most severe drug-resistant serotype. MLST showed that there was correlation between serotypes and genotypes in most Salmonella isolates, except S. Pullorum, which showed complicated genetic diversity firstly. These results provide important epidemiological information for us to control Salmonella in chickens.
Similar content being viewed by others
Background
Salmonella is an important zoonotic pathogen that causes infectious diseases in animals and humans [1] . Every year, Salmonella infection causes not only decreased production performance and even death of poultry, but also contamination of the human food chain, leading to serious economic losses in the poultry industry, as well as being a threat to public health [2]. Although various prevention and control measures, including eradication programs and vaccine and drug use, have been carried out [3], Salmonella infection is still one of the most important problems worldwide.
Currently, based on the difference in O, H and Vi antigens, > 2600 different Salmonella serotypes have been identified [4]. It is interesting that, although their genomes are similar, the ranges of host in different serotypes are discrepant. Salmonella enterica serovar Gallinarum is a host-specific pathogen that only infects birds. It has two biovars, Salmonella enterica serovar Gallinarum biovar Gallinarum (S. Gallinarum) and Salmonella enterica serovar Gallinarum biovar Pullorum (S. Pullorum), which cause fowl typhoid and pullorum disease in poultry, respectively [5], leading to great economic losses in the poultry industry, especially in China. Salmonella enterica serovar Typhimurium and Salmonella enterica serovar Enteritidis are the most common serotypes of Salmonella, which can infect broad hosts, including humans and birds. S. Typhimurium causes a typhoid-like systemic illness and S. Enteritidis is the main cause of acute gastroenteritis in humans [6]. These serotypes of Salmonella are considered to be the most important food-borne pathogens worldwide. Other serotypes such as Salmonella enterica serovar Heidelberg, Salmonella enterica serovar Kentucky and Salmonella enterica serovar Newport are also reported in chickens [7]. Because of the differences in pathogenicity and hosts among different serotypes, to understand the dominant serotypes, virulence factors and genetic characteristics of prevailing strains will help us further develop control strategies in the poultry industry.
Antimicrobial resistance and its spread are also a serious problem caused by Salmonella. Although the Chinese government is promoting reduction of antibiotic use currently, antimicrobial drugs remain one of the important options for Salmonella control [8], which leads to an increase of multiple drug-resistant bacteria worldwide. From 1970 to 2010 in China, the resistance rates to antimicrobials, such as ampicillin, gentamicin, streptomycin, tetracycline, and chloramphenicol, were increasing, and now remain at high levels in S. Pullorum [9]. Ampicillin resistance in Salmonella has been recognized as the most serious problem in some Asian countries, such as in Bangladesh (68.4%) and Vietnam (80.4%) [10]. The extensive use of antimicrobials in animal production has resulted in contamination by multidrug-resistant Salmonella in food animals [11]. Recently, a series of resistance genes for human antimicrobials, such as mcr-1 and tetX3/4, was discovered in zoonotic bacteria [12]. This is a major public health concern because animal-derived antibiotic-resistant bacteria can be transmitted to humans [13].
Poultry is the most important host of Salmonella, so assessing the distribution of Salmonella in poultry has become important for better prevention and control of its infection. In this study, we investigated the main serotypes of Salmonella isolated from sick and healthy-looking chickens, and assessed their genetic relationship among our isolates, presence of potential virulence factors and antimicrobial-resistance profiles.
Results
Serotype identification of Salmonella isolates from chickens
A total of 84 strains, including 41 from sick chickens and 43 from healthy-looking chickens were identified as Salmonella, and further serotyped based on agglutination tests and PCR tests. Salmonella serogroup D, including S. Enteritidis, S. Pullorum and S. Gallinarum accounted for 84.76% of our isolates, and among them, S. Pullorum was the most common type in both sick chickens (n = 18) and healthy-looking chickens (n = 20) (Table 1). In addition, 11 isolates (2 from healthy-looking chickens and 9 from sick chickens) were identified as S. Typhimurium, and 16 isolates (5 from healthy-looking chickens and 11 from sick chickens) were identified as S. Gallinarum. It is interesting that S. Enteritidis (n = 14) was only found in healthy-looking chickens.
Antimicrobial susceptibilities of different serotypes of Salmonella isolates
The susceptibility of Salmonella isolates to 10 antimicrobials was tested using MIC assays. The isolates showed high resistance rates to ampicillin (97.6%), tetracycline (58.3%), and colistin (51.2%), and lower resistance rates for gentamicin (1.2%), tigecycline (4.8%), ciprofloxacin (6.0%), cefotaxime (7.1%), and meropenem (7.1%) (Table 2). Among these isolates, higher resistance rates to colistin (64.3%), meropenem (35.7%) and ciprofloxacin (28.6%) were observed in S. Enteritidis, which can infect humans, while cefotaxime resistance was only observed in S. Gallinarum (25%) and S. Pullorum (5.3%), which only infect birds. The isolates showed 30 different antimicrobial-resistance patterns, and only one S. Gallinarum strain was sensitive to all the antimicrobials tested (Table 3). Forty-nine isolates (59.5%) were resistant to ≥3 antimicrobials in different combinations. Resistance to ampicillin, tetracycline, and colistin was the most common multidrug-resistance profile, accounting for 9.6%. One S. Enteritidis isolate was resistant to seven antimicrobials, and six of 14 (42.9%) S. Enteritidis isolates were resistant to more than five antimicrobials (Table 3). S. Enteritidis showed highest resistance rates among different serotypes of isolates in six of 10 tested antimicrobials, including ampicillin, meropenem, tetracycline, ciprofloxacin, trimethoprim-sulfamethoxazole, and colistin (Table 2).
Genetic diversity of Salmonella isolates
The Salmonella isolates were classified into four clades based on the MLST genotypes (Fig. 1). The allele numbers and the sequences of housekeeping genes in each ST are shown in Additional file 1. All of the S. Gallinarum and S. Enteritidis isolates were clustered in clade 1, and among them, 15 of 16 S. Gallinarum isolates belong to ST-1747, which is not reported currently, while the remaining one S. Gallinarum and all the S. Enteritidis isolates belonged to ST-11. All the S. Typhimurium isolates were clustered in clade 3, and belonged to ST-128, ST-1544 and ST-19. In S. Pullorum, 71.1% isolates belonged to the dominant genotype ST-92, while another six ST types were identified, including ST-2151, ST-11, ST-1747, ST128, ST-1544 and ST-99, which showed the genetic diversity of S. Pullorum. Among these S. Pullorum isolates, three isolates belonged to ST-99, ST-128 and ST-1544 were clustered in clade 3.
Distribution of virulence-associated genes in different serotypes of Salmonella isolates
As shown in Table 4, cdtB was not found in all isolates, while the remaining of detected virulence-associated genes were found in all serogroup D Salmonella (S. Enteritidis, S. Pullorum and S. Gallinarum). In contrast, sefA and sopE were absent in one and two S. Typhimurium isolates, respectively, and sodC1 and sopE were absent in one other serotype of Salmonella (JC180), which was the only isolates in clade 3 in this study.
Discussion
The distribution of Salmonella serovars in poultry differs among countries and regions. In India, S. Typhimurium, S. Gallinarum and S. Enteritidis were the most prevalent serovars, accounting for 96.2% of isolates [14]. In Egypt, S. Enteritidis and S. Typhimurium were the most commonly identified serotypes recovered from broiler chickens and retail shops [15]. In contrast, an investigation from Japan showed that only Salmonella enterica serovar Infantis, Salmonella enterica serovar Manhattan and Salmonella enterica serovar Schwarzengrund were present in cecal samples of broiler chickens [16], which was different from China. In our study, the main types of isolated Salmonella were S. Pullorum, S. Gallinarum, S. Enteritidis, and S. Typhimurium, and among them, S. Pullorum was the most common type in both healthy-looking and sick chickens. Although eradication programs for S. Pullorum from breeding birds are underway in most of the poultry farms, pullorum disease is still one of the most severe diseases in poultry. S. Pullorum can cause acute sepsis in chicks, mostly infects young chicks within 20 days of age, and the morbidity and mortality are high [17]. In contrast, carrier state without obvious symptoms frequently occurs in adult chickens, which become an important source of transmission [18]. It is well-known that the transmission of S. Pullorum occurs both horizontally and vertically [19], and the strong and multipath transmission might play an important roles in its spread. It is particularly interesting that S. Enteritidis was present in healthy-looking chickens, which suggested that the pathogenicity of the prevalent strains was weak in chickens. However, S. Enteritidis colonizes the gastrointestinal tract of poultry, resulting in no clinical symptoms in chickens, but causing gastroenteritis in humans [20]. It has become one of the most commonly reported causes of foodborne illness in humans [21]. Although a vaccine is used to control S. Enteritidis in the US and Europe [22], there is no good measure for its control in chickens in China. High isolation rate of S. Enteritidis during the slaughtering process has been reported in China [23]. We think that carriage in healthy-looking chickens means that there is a high risk of S. Enteritidis entering the food processing stage, which highlights its public health threat.
It is thought that animal breeding is an important source of resistant pathogens. Therefore, antimicrobial resistance is a serious problem in the poultry industry as well as a threat to public health. High resistance rates to ampicillin (97.6%), tetracycline (58.3%) and colistin (51.2%) were observed, and 59.5% of isolates were resistant to three or more antimicrobials in different combinations. Therefore, multidrug resistance limits the choices of treatment of Salmonella infection in chickens. High resistance rates to ampicillin and tetracycline have been found in isolates from poultry or poultry products in many countries, such as the US (ampicillin, 85%; tetracycline, 35%) [24], Malaysia (ampicillin, 89.5%; tetracycline, 85.1%) [25], and Egypt (ampicillin, 86.7%; tetracycline, 40.0%) [26], as well as in patient with gastroenteritis [27]. However, serious resistance to colistin is seen in only a few countries, including China [28], but colistin used to be considered the last choice for treatment for Gram-negative bacterial infection [29]. What is more important, S. Enteritidis which can infect humans, is the most severe drug-resistant serotype. Higher resistance rates to colistin, meropenem and ciprofloxacin, which are often used for human treatment, were observed in this serotype. As mentioned, S. Enteritidis is carried in healthy-looking chickens and has a high risk of entering the food chain, so its resistance is closely related to human health. Therefore, this serious problem is not only harmful for poultry industry but also public health.
In this study, the genetic relationship of 84 Salmonella isolates was determined by MLST. All of the S. Enteritidis and S. Gallinarum isolates were clustered in the first subgroup of clade 1, while all the S. Typhimurium isolates were clustered in clade 2, suggesting a strong correlation between STs and serotypes. However, S. Pullorum isolates spread in clades 1, 2 and 3, which showed complex genetic diversity. Besides previously reported ST-92, which is the dominant ST of S. Pullorum, and ST-2151 [30], some STs, including ST-1747, ST-11, ST-99, ST-1544 and ST-128, were firstly identified in S. Pullorum isolates. Even three strains (ST-99, ST-1544 and ST-128) showed high genetic similarity to S. Typhimurium. As in previous studies, S. Gallinarum and S. Pullorum, which share the same O antigens 1, 9 and 12, were a direct descendant of S. Enteritidis after host adaption [31]. Our results suggested that, there was correlation between serotypes and genotypes in most Salmonella isolates. However, S. Pullorum has evolved toward diversity with long-term colonization in birds, and this might also be a reason for the high prevalence of S. Pullorum among poultry in China.
Although the genetic background of these Salmonella serotypes was similar, the host specificity and pathogenicity differed among serotypes. S. Typhimurium and S. Enteritidis had wide host ranges, and the former caused a typhoid-like systemic illness, while the latter was the main cause of acute gastroenteritis in humans. In contrast, S. Pullorum and S. Gallinarum were bird specific, and showed different pathological symptoms. We detected the prevalence of 27 virulence-associated genes, which were involved in invasion and intracellular survival, among our isolates. The pathogenicity of Salmonella is mainly reflected in its ability to invade non-phagocytic cells, survive in phagocytic cells, and replicate and proliferate in phagocytic cells, all of which are closely related to the virulence factors encoded in the Salmonella pathogenicity island (SPI) [32]. For example, inactivation of sipB in Salmonella enterica serotype Dublin strongly reduces fluid secretion and inflammation [33], and deletion of prgH result in strongly reduced virulence of S. Typhimurium [34]. Knock out of sodCI or spv genes reduces persistence of S. Enteritidis in mice [35]. We found that these 27 virulence-associated genes were relatively conserved in our Salmonella isolates, especially in serogroup D Salmonella, and suggested that these there was no direct correlation between serotypes and the distribution of these virulence genes. The genomic sequences of different serotypes revealed that host-restricted Salmonella had undergone more extensive degradation than host promiscuous Salmonella [36]. According to their study, two of our detected genes, lpfC and shdA, which were involved in host colonization, were identified as pseudogenes in S. Pullorum and S. Gallinarum [31]. In addition, removed metabolic pathways due to reduced genomes might be a more important factor for niche adaption processes [31], rather than prevalence of virulence-associated genes. Therefore, we think although these 27 genes were important for the pathogenicity of Salmonella, they were not the only key factors for their virulence and host ranges.
Conclusion
Salmonella serogroup D was the major subgroup, and S. Pullorum was the most common type in sick and healthy-looking chickens in central China. Drug resistance assays showed high resistance rates to ampicillin, tetracycline and colistin, and among them, S. Enteritidis was the most severe drug-resistant serotype, which showed higher resistance rates to colistin, meropenem and ciprofloxacin. MLST showed that there was correlation between serotypes and genotypes in most Salmonella isolates, except S. Pullorum, which showed complicated genetic diversity. These results provide important epidemiological information for control of Salmonella in chickens.
Methods
Identification and serotyping of Salmonella isolates
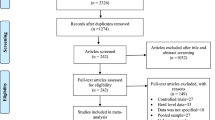
During 2013–2018, there were 84 strains of Salmonella that were mainly isolated in central China (Henan, Jiangsu and Hubei). Sick chickens were euthanized with an intramuscular injection of sodium pentobarbital (100 mg/kg bodyweight) according to Hubei Province Laboratory Animal Management Regulations – 2005. Samples were isolated from brain, lung, spleen and liver of dead or sick chickens on poultry farms, and isolated from anal swabs of healthy-looking chickens on poultry farms or at poultry markets (Fig. 1). To isolate the Salmonella, collected disease samples or anal swabs were directly transported to the laboratory. Each sample was inoculated into 225 mL of buffered peptone water (BPW; Land Bridge, China). After 16 h incubation at 37 °C, 0.1 mL and 10 mL of BPW were transferred into 10 mL of tetrathionate broth and 100 mL of selenite broth, respectively. Subcultures were spread on xylose-lysine-deoxycholate (XLD; Land Bridge, China) agar, xylose-lysine-tergitol 4 (XLT4; Land Bridge, China) agar, and Hektoen (HE; Hopebio, China) agar, and incubated at 37 °C for 24 h. The typical Salmonella colonies were confirmed by PCR amplification of the hut gene, and the primers were as follows, hut-F, 5′-ATGTTGTCCTGCCCCTGGTAAGAGA-3′, hut-R, 5′-ACTGGCGTTATCCCTTTCTCTGCTG-3′ [37]. Serotypes were determined by a slide agglutination test with O-antigen antiserum and a tube agglutination test with H-antigen antiserum, and confirmed by quadruplex PCR analysis as previously reported [38]. After the experiment, the Salmonella-positive chickens were euthanized and underwent harmless treatment based on the regulations from Hubei Provincial Animal Care and Use Committee.
Antimicrobial resistance profiles
The minimum inhibitory concentrations (MICs) of gentamicin, meropenem, ampicillin, cefotaxime, colistin, tetracycline, ciprofloxacin, chloramphenicol, trimethoprim-sulfamethoxazole and tigecycline were determined according to the Clinical and Laboratory Standards Institute [39]. Escherichia coli ATCC 25922 was used for quality control. Isolates were classified as susceptible, intermediate or resistant according to their MICs.
MLST and phylogenetic tree
To analyze the genetic diversity, MLST was carried out. The isolates were grown overnight in Luria-Bertani medium at 37 °C, and DNA was extracted using MiniBEST Universal Genomic DNA Extraction Kit (TaKaRa, Dalian, China). The following seven housekeeping genes, aroC, dnaN, hemD, hisD, purE, sucA, and thrA [40], in each tested isolate were amplified by PCR and the PCR products were sequenced by bi-directional DNA sequencing. The obtained sequences were analyzed using salmonella MLST database (http://enterobase.warwick.ac.uk/species/senterica/allele_st_search), and the allele numbers and sequence types (STs) were assigned. The phylogenetic tree was constructed using MEGA cluster analysis.
Detection of virulence-associated genes
Considering the contribution of virulence genes to the invasiveness and pathogenicity of Salmonella, a total of 27 virulence-associated genes were screened by PCR. Twenty seven virulence-associated genes, including sodC1, spvC, spvB, spiAi, pagC, cdtB, msgA, invA, sipB, prgH, spaN, orgA, tolC, iroN, sitC, lpfC, sifA, sopB, sefA, pipA, ttrC, misL, siiE, mgtB, spi4D, shdA, and sopE, were detected by PCR tests. The primers and amplification conditions were as previously described [41]. PCR was performed in a GeneAmp PCR System 2720 (Applied Biosystems, Singapore). The PCR cycling consisted of 35 cycles of 30 s at 94 °C, 30 s at 55 °C and 1 min at 72 °C and resulting amplification products were separated by electrophoresis in 2% agarose gel, stained with ethidium bromide and visualized under UV light.
Availability of data and materials
The DNA sequences generated and/or analysed during the current study are available in the GenBank repository, Accession: from MT799111 to MT799173. The others datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- S. Enteritidis:
-
Salmonella enterica serovar Enteritidis
- S. Typhimurium:
-
Salmonella enterica serovar Typhimurium
- S. Gallinarum:
-
Salmonella enterica serovar Gallinarum biovar Gallinarum
- S. Pullorum:
-
Salmonella enterica serovar Gallinarum biovar Pullorum
- MLST:
-
Multilocus Sequence Typing
- STs:
-
Sequence types
- MICs:
-
Minimum inhibitory concentrations
References
Li Q, Wang X, Xia J, Yuan Y, Yin C, Xu L, Li Y, Jiao X. Salmonella-containing vacuole development in avian cells and characteristic of cigR in Salmonella enterica serovar Pullorum replication within macrophages. Vet Microbiol. 2018;223:65–71.
Sylejmani D, Musliu A, Ramadani N, Sparagano O, Hamidi A. Associations between the level of biosecurity and occurrence of Dermanyssus gallinae and Salmonella spp. in layer farms. Avian Dis. 2016;60(2):454–9.
Zhang-Barber L, Turner AK, Barrow PA. Vaccination for control of Salmonella in poultry. Vaccine. 1999;17(20–21):2538–45.
Hendriksen RS, Vieira AR, Karlsmose S, DMA LFW, Jensen AB, Wegener HC, Aarestrup FM. Global monitoring of Salmonella Serovar distribution from the World Health Organization global foodborne infections network country data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis. 2011;8(8):887–900.
Soria MC, Soria MA, Bueno DJ. Comparison of 2 culture methods and PCR assays for Salmonella detection in poultry feces. Poult Sci. 2012;91(3):616–26.
Vugia DJ, Samuel M, Farley MM, Marcus R, Shiferaw B, Shallow S, Smith K, Angulo FJ. Invasive Salmonella infections in the United States, FoodNet, 1996-1999: incidence, serotype distribution, and outcome. Clin Infect Dis. 2004;38(Suppl 3):S149–56.
Long M, Lai H, Deng W, Zhou K, Li B, Liu S, Fan L, Wang H, Zou L. Disinfectant susceptibility of different Salmonella serotypes isolated from chicken and egg production chains. J Appl Microbiol. 2016;121(3):672–81.
Zhang QQ, Ying GG, Pan CG, Liu YS, Zhao JL. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environment Sci Technol. 2015;49(11):6772–82.
Gong J, Kelly P, Wang C. Prevalence and antimicrobial resistance of Salmonella enterica Serovar Indiana in China (1984-2016). Zoonoses Public Health. 2017;64(4):239–51.
Chiou CS, Lauderdale TL, Phung DC, Watanabe H, Kuo JC, Wang PJ, Liu YY, Liang SY, Chen PC. Antimicrobial resistance in Salmonella enterica Serovar Typhi isolates from Bangladesh, Indonesia, Taiwan, and Vietnam. Antimicrob Agents Chemother. 2014;58(11):6501–7.
Chuanchuen R, Ajariyakhajorn K, Koowatananukul C, Wannaprasat W, Khemtong S, Samngamnim S. Antimicrobial resistance and virulence genes in Salmonella enterica isolates from dairy cows. Foodborne Pathog Dis. 2010;7(1):63–9.
He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, Ke Y, Ji Q, Wei R, Liu Z, et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol. 2019;4(9):1450–6.
Khemtong S, Chuanchuen R. Class 1 integrons and Salmonella genomic island 1 among Salmonella enterica isolated from poultry and swine. Microb Drug Resist. 2008;14(1):65–70.
Kumar Y, Singh V, Kumar G, Gupta NK, Tahlan AK. Serovar diversity of Salmonella among poultry. Indian J Med Res. 2019;150(1):92–5.
Elkenany R, Elsayed MM, Zakaria AI, El-Sayed SA, Rizk MA. Antimicrobial resistance profiles and virulence genotyping of Salmonella enterica serovars recovered from broiler chickens and chicken carcasses in Egypt. BMC Vet Res. 2019;15(1):124.
Duc VM, Nakamoto Y, Fujiwara A, Toyofuku H, Obi T, Chuma T. Prevalence of Salmonella in broiler chickens in Kagoshima, Japan in 2009 to 2012 and the relationship between serovars changing and antimicrobial resistance. BMC Vet Res. 2019;15(1):108.
Barrow PA, Freitas Neto OC. Pullorum disease and fowl typhoid--new thoughts on old diseases: a review. Avian Pathol. 2011;40(1):1–13.
Wigley P, Berchieri A Jr, Page KL, Smith AL, Barrow PA. Salmonella enterica serovar Pullorum persists in splenic macrophages and in the reproductive tract during persistent, disease-free carriage in chickens. Infect Immun. 2001;69(12):7873–9.
Liu GR, Rahn A, Liu WQ, Sanderson KE, Johnston RN, Liu SL. The evolving genome of Salmonella enterica serovar Pullorum. J Bacteriol. 2002;184(10):2626–33.
Paiva JB, Penha Filho RA, Junior AB, Lemos MV. Requirement for cobalamin by Salmonella enterica serovars Typhimurium, Pullorum, Gallinarum and Enteritidis during infection in chickens. Brazilian J Microbiol. 2011;42(4):1409–18.
Chousalkar K, Gast R, Martelli F, Pande V. Review of egg-related salmonellosis and reduction strategies in United States, Australia, United Kingdom and New Zealand. Crit Rev Microbiol. 2018;44(3):290–303.
Methner U. Immunisation of chickens with live Salmonella vaccines - role of booster vaccination. Vaccine. 2018;36(21):2973–7.
Zhu Y, Lai H, Zou L, Yin S, Wang C, Han X, Xia X, Hu K, He L, Zhou K, et al. Antimicrobial resistance and resistance genes in Salmonella strains isolated from broiler chickens along the slaughtering process in China. Int J Food Microbiol. 2017;259:43–51.
Gad AH, Abo-Shama UH, Harclerode KK, Fakhr MK. Prevalence, serotyping, molecular typing, and antimicrobial resistance of Salmonella isolated from conventional and organic retail ground poultry. Front Microbiol. 2018;9:2653.
Chuah LO, Shamila Syuhada AK, Mohamad Suhaimi I, Farah Hanim T, Rusul G. Genetic relatedness, antimicrobial resistance and biofilm formation of Salmonella isolated from naturally contaminated poultry and their processing environment in northern Malaysia. Food Res Int. 2018;105:743–51.
Moawad AA, Hotzel H, Awad O, Tomaso H, Neubauer H, Hafez HM, El-Adawy H. Occurrence of Salmonella enterica and Escherichia coli in raw chicken and beef meat in northern Egypt and dissemination of their antibiotic resistance markers. Gut Pathog. 2017;9:57.
Luo Y, Yi W, Yao Y, Zhu N, Qin P. Characteristic diversity and antimicrobial resistance of Salmonella from gastroenteritis. J Infect Chemother. 2018;24(4):251–5.
Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–8.
Hu Y, Liu F, Lin IY, Gao GF, Zhu B. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(2):146–7.
Guo X, Wang H, Cheng Y, Zhang W, Luo Q, Wen G, Wang G, Shao H, Zhang T. Quinolone resistance phenotype and genetic characterization of Salmonella enterica serovar Pullorum isolates in China, during 2011 to 2016. BMC Microbiol. 2018;18(1):225.
Feng Y, Johnston RN, Liu GR, Liu SL. Genomic comparison between Salmonella Gallinarum and Pullorum: differential pseudogene formation under common host restriction. PLoS One. 2013;8(3):e59427.
Dandekar T, Fieselmann A, Fischer E, Popp J, Hensel M, Noster J. Salmonella-how a metabolic generalist adopts an intracellular lifestyle during infection. Front Cell Infect Microbiol. 2014;4:191.
G EE, W MW, R R, M PB, W PR, H S. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol. 1997;25(5):903–12.
T RM, A LG, F TA. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun. 1999;67(9):4879–85.
Karasova D, Havlickova H, Sisak F, Rychlik I. Deletion of sodCI and spvBC in Salmonella enterica serovar Enteritidis reduced its virulence to the natural virulence of serovars Agona, Hadar and Infantis for mice but not for chickens early after infection. Vet Microbiol. 2009;139(3–4):304–9.
Thomson NR, Clayton DJ, Windhorst D, Vernikos G, Davidson S, Churcher C, Quail MA, Stevens M, Jones MA, Watson M, et al. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 2008;18(10):1624–37.
Alzwghaibi AB, Yahyaraeyat R, Fasaei BN, Langeroudi AG, Salehi TZ. Rapid molecular identification and differentiation of common Salmonella serovars isolated from poultry, domestic animals and foodstuff using multiplex PCR assay. Arch Microbiol. 2018;200(7):1009–16.
Xiong D, Song L, Tao J, Zheng H, Zhou Z, Geng S, Pan Z, Jiao X. An efficient multiplex PCR-based assay as a novel tool for accurate inter-Serovar discrimination of Salmonella Enteritidis, S Pullorum/Gallinarum and S Dublin. Front Microbiol. 2017;8:420.
CLSI: Performance Standards for Antimicrobial Susceptibility Testing M100S, 26th Edition. 2017.
Maiden MC. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–88.
Skyberg JA, Logue CM, Nolan LK. Virulence genotyping of Salmonella spp. with multiplex PCR. Avian Dis. 2006;50(1):77–81.
Acknowledgements
Not applicable.
Funding
This work was supported by National Key Research and Development Program of China (2016YFD0501305), China Agriculture Research System (CARS-41-G13) and Technical Innovation Project of Hubei Province (2018ABA108). The funders had no role in study design, data collection, analysis and interpretation, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
XW, GW and TZ participated in the conception and design of the study. TL, YC, HW and XG contributed to the collection of samples. XW, HW and FL performed the farm and laboratory work. XW, ZP and TZ analyzed the data and wrote the manuscript. QL and HS contributed to the analysis and helped in the manuscript discussion. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All animal studies were conducted in strict accordance with the guidelines of animal welfare of World Organization for Animal Health. The experiments were approved by the Ethics Committee of Hubei Academy of Agricultural Sciences according to Hubei Province Laboratory Animal Management Regulations – 2005. The animals used in this study were derived from commercial sources, and the owners’ consent was not required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, X., Wang, H., Li, T. et al. Characterization of Salmonella spp. isolated from chickens in Central China. BMC Vet Res 16, 299 (2020). https://doi.org/10.1186/s12917-020-02513-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-020-02513-1