Abstract
Background
A microbiological diagnosis is essential to better target antimicrobial treatment, control and prevention of respiratory tract infections in cattle. Under field conditions, non-endoscopic broncho-alveolar lavage (nBAL) samples are increasingly collected. To what extent the highly variable turnaround time and storage temperatures between sampling and cultivation affect the isolation rate of bacterial pathogens is unknown. Therefore, the objective of this experimental study was to determine the effect of different storage temperatures (0 °C, 8 °C, 23 °C and 36 °C) and times (0,2,4,6,8,24,48 h) on the isolation rate and concentration of Pasteurellaceae in nBAL samples from clinically affected animals.
Results
At a storage temperature temperature of 36 °C isolation rates of Mannheimia haemolytica and Pasteurella multocida were significantly reduced 6 h and 48 h after sampling, respectively. At room temperature (23 °C), a decrease in M. haemolytica and P. multocida isolation rate was noticed, starting at 24 and 48 h after sampling, respectively, but only significant for P. multocida at 48 h. The presence of microbial contamination negatively affected the isolation of P. multocida in clinical nBAL samples, but not of M. haemolytica.
Conclusion
Optimal M. haemolytica and P. multocida isolation rates from clinical nBAL samples are obtained after storage at 0 °C or 8 °C, provided that the sample is cultivated within 24 h after sampling. The maximum period a sample can be stored without an effect on the M. haemolytica and P. multocida isolation success varies and is dependent on the storage temperature and the degree of microbial contamination.
Similar content being viewed by others
Background
Respiratory tract infections (bovine respiratory disease (BRD)) have a major impact on farm economics and animal welfare [1]. Furthermore, they are the main indication for antimicrobial use in calves [2]. In order to rationalize antimicrobial use, diagnostic techniques need to be optimized. Non-endoscopic bronchoalveolar lavage (nBAL) is a practical and economical technique, increasingly used in Western European countries to sample the lower airways of cattle [3]. Although this technique obtains more pure cultures compared to a deep nasopharyngeal swab, microbial sample contamination can occur, partly depending on the experience of the veterinarian [3]. In order to minimize microbial contamination and bacterial overgrowth, cultivation of samples needs to be performed as soon as possible after sampling. However, due to the centralization of veterinary laboratories and the limited operating hours of these laboratories (not 24/7 as in some human clinics), the turnaround time between sampling and cultivating of the samples can take 24 h to even several days. Optimal storage conditions, supporting survival of causal pathogens and limiting growth of contaminants, are needed to obtain relevant bacterial analysis results [4]. False negative or irrelevant results may lead to therapy failure, resulting in increased antimicrobial use, antimicrobial resistance and mortality. Although storage conditions of clinical samples in the field are important, only few studies addressed this subject. Two studies are available comprising the effects of long term survival of Pasteurellaceae, namely in swabs from bears [5] or ovine and bovine tracheobronchial washings [6]. In the latter experiments, however, sterile lung fluids were spiked, and therefore possible contaminant effects were not taken into account. To what extent nBAL field samples can be stored until analysis, without influencing the isolation rate of clinically import pathogens, is currently unknown. Therefore, the objective of the present study was to determine the effect of storage temperature and duration on the isolation rate of Pasteurellaceae from bovine nBAL field samples.
Results
Animals that met the inclusion criteria aged 1 week to 7 months. In total, 13 nBAL samples were eligable, of which the initial culture results at T0 showed 6 dominant cultures with 1 clinically relevant pathogen (4 M. haemolytica, 2 P. multocida) and 7 mixed cultures with both M. haemolytica and P. multocida (3 pure cultures, meaning the presence of 1 bacterial species on the agar plate with more than 2 colonies, containing only M. haemolytica and P. multocida and 4 dominant cultures with also some contaminants present). In total, M. haemolytica was isolated from 11 samples (84.6%) with an average log concentration of 4.0 at T0, and P. multocida from 9 samples (69.2%) with an average log concentration of 3.7 at T0. Trueperella pyogenes was isolated from one sample and Moraxella bovis from two samples at T0, both in mixed cultures. H. somni was not isolated. Contaminants present in the dominant cultures were Streptococcus spp., Staphylococcus spp., Bacillus spp., Escherichia coli and Rothia nasimurium with an average log concentration of 2.8 at T0.
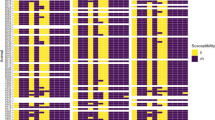
The effect of the various temperatures and storage periods after sampling on the number of positive samples for M. haemolytica is presented in Fig. 1. In general, the number of samples from which M. haemolytica could be isolated, decreased gradually over time. The higher the storage temperature, the earlier the number of positive samples started to decrease, i.e. at 2 h of storage at 36 °C, at 24 h of storage at 23 °C and at 48 h of storage at 0 °C and 8 °C. During storage at 0 °C and 8 °C, the number of positive samples remained stable up to 24 h after sampling, varying between 10/11 and 11/11 in this period, whereas at 23 °C, only 7/11 samples remained positive after 24 h of storage. When samples were stored at 36 °C for 48 h, M. haemolytica could be isolated from only 1 sample, while this was 4/11 for 23 °C and 8/11 for both 0 °C and 8 °C (Fig. 1). Significance was only reached after 6 h of storage at 36 °C compared to lower temperatures.
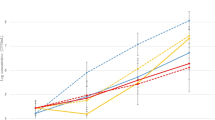
An overall slight decrease in M. haemolytica concentration occurred over time (Fig. 2). At a storage temperature of 36 °C, the average log concentration of M. haemolytica decreased after 2 h of storage from 4 to 3.5 and remained stable until 48 h after sampling. No statistical significant difference was seen between the different temperatures and the time after sampling.
The effect of storage temperature and time on the number of samples from which P. multocida could be isolated is presented in Fig. 3. A significant difference of isolation is only seen at 48 h of storage at a temperature of 0 °C or 8 °C, 23 °C and 36 °C. A decline in the number of positive samples from which P. multocida was isolated was observed throughout the experiment for storage at 36 °C, where only 4/9 positive samples were retrieved at 24 h of storage and no positive samples could be retrieved at 48 h (Fig. 3). This decline in the number of positive samples was due to both contaminant overgrowth and a decreased viability. This decreased viability of P. multocida was noticed at a storage temperature of 36 °C starting from 24 h after sampling and at a storage temperature of 23 °C starting from 48 h after sampling. When samples were maintained for 48 h at 23 °C, only 3/9 samples were found positive. At a storage temperature of 0 °C or 8 °C, the number of positive samples remained stable until 24 h after sampling, with an isolation rate of 8/9 to 9/9. At 48 h of storage at a temperature of 0 °C, 1 initially P. multocida positive sample was negative due to decreased viability. When stored at 8 °C for 48 h, 2 initially P. multocida positive samples were negative, one due to decreased viability and one due to contaminant overgrowth.
Compared with M. haemolytica, the average concentration of P. multocida remained more stable until 24 h of storage at a temperature of 0 °C, 8 °C and 23 °C (Fig. 4). At a storage temperature of 36 °C, the P. multocida concentration slightly decreased starting from 6 h after sampling, with a concentration below detection limit at 48 h after sampling (Fig. 4). Accordingly, no positive samples of P. multocida were retrieved after storage at 36 °C for 48 h, even in the absence of contamination overgrowth. No statistical significant difference was seen between the different temperatures and the time after sampling.
From the 13 samples collected, 10 samples contained microbial contamination at T0. Of the 3 initial samples that were not contaminated at T0, 1 sample showed microbial contamination starting from 2 h after sampling at a temperature of 36 °C. The other two initially negative samples showed sporadically microbial contamination with a concentration close to the detection limit. Results of the influence of bacterial contamination on the isolation rate of M. haemolytica and P. multocida in the clinical nBAL samples are presented in Table 1. A statistically significant negative association was seen between the presence of contaminants and the presence of P. multocida. An odds ratio of 0.32 was obtained for P. multocida (P = 0.04), meaning that the presence of contaminants reduced the odds of isolating P. multocida. In contrast, for M. haemolytica no significant effect of the presence of contaminants on isolation rates could be evidenced (P = 0.70). An average initial contaminants log concentration of 2.8 CFU/mL was observed (Fig. 5). This concentration remained stable during the first 8 h of storage, regardless of storage temperature. However, after 24 h of storage at 36 °C, the average contaminants log concentration increased to 4.4. After 48 h of storage, an average contaminants log concentration of 4.7 and 5.1 was reached for storage at 23 °C and 36 °C, respectively (Fig. 5).
Discussion
This study describes the effects of different storage temperatures and times on the isolation rate and concentration of Pasteurellaceae from nBAL samples. When nBAL samples were stored at a temperature of 0 °C or 8 °C, high isolation rates of P. multocida and M. haemolytica were obtained until 24 h of storage. Since storage at 0 °C has low feasibility in routine practice, storage of clinical nBAL samples in a refrigerator is a readily available alternative for most practitioners. Also in previous studies low temperatures are recommended for storage [7, 8].
Even though there was no statistically significant decrease in the isolation rate after 24 h of storage at room temperature (23 °C) for both P. multocida and M. haemolytica, a probably relevant decrease in M. haemolytica isolation rate (from 11/11 to 7/11) was observed. The reasons for this decreased isolation rate were both a decreased viability (2/4 samples) and microbial contamination (2/4 samples). According to Tano et al. [9], clinically important bacteria can stay viable for 24 h at room temperature, but not in polymicrobial samples. However, these samples were spiked with high pathogen concentrations (106 CFU/mL). When lower concentrations, comparable with concentrations obtained in the present study, were used (104 CFU/mL and 105 CFU/mL), results in viability varied. In another study M. haemolytica remained viable for a long period of up to 156 days [6]. However in that studies samples were also spiked with a high concentration (106 – 107 CFU/mL) and no bacterial contamination was present. These results stress the importance of a high initial pathogen concentration and avoiding microbial contamination during and after the sampling procedure, in order to maximize the probability to isolate Pasteurellaceae.
When samples were stored at 36 °C, the isolation rate started to decline already 2 and 24 h after sampling for M. haemolytica and P. multocida, respectively. When samples were maintained for 48 h at these temperatures, isolating clinically relevant pathogens was rare or no longer possible. This was mainly, but not exclusively, due to the increased concentration of contaminants, leading to uninterpretable samples when a concentration of ≥1 × 106 CFU/mL of contaminants was reached. These results stress the importance of not leaving samples above room temperature, for example in a closed car or in a tropical environment.
The concentration of M. haemolytica slightly decreased over time independent of the storage temperature, though not statistically significant, while for P. multocida, this concentration remained more stable. Currently, no information is available on the survival rate of latter 2 bacteria in nBAL samples. One recent study describes the survival rate and density range of Pasteurellaceae in the nasopharyngeal microbiota in healthy calves [10]. This study showed a longer duration of carriage in the nose and higher concentration for P. multocida compared to H. somni, however rates of M. haemolytica were too low for meaningful survival modelling. Retaining high concentrations of relevant bacteria in clinical samples can be an added value to diagnostics, both for clinical interpretation as for direct detection methods using for example matrix-assisted laser desorption/ionization-time-of- flight mass spectrometry (MALDI-TOF MS) [11]. A higher negative association between the presence of contaminants and the isolation rate of P. multocida was found compared to M. haemolytica. This might be partially due to the fact that the average initial load of M. haemolytica in the samples was higher compared with the initial P. multocida load. Contaminant overgrowth might therefore negatively impact P. multocida isolation rate more quickly than the M. haemolytica isolation rate. A previous study showed that the growth of M. haemolytica can be inhibited by contaminants like Escherichia coli rather than Staphylococcus spp. or Streptococcus spp. [12]. To what extent different bacterial contaminants had an inhibitory effect on P. multocida and/or M. haemolytica in this study is unclear, considering the limited number of samples and since different bacterial contaminants were often combined at different concentrations within one clinical sample.
A limitation of the current study is the limited sample size. When using 8 positive samples per test group, only 60% difference in isolation rate could be detected. One of the reasons for the limited number of used samples are the strict inclusion and exclusion criteria applied. Indeed, only samples obtained from untreated calves, well-characterized as clinically affected at the level of the lower respiratory tract were included. In addition, only samples from which clinically relevant bacteria could be isolated at T0 were included in the experiment, resulting in the exclusion of various samples. Nevertheless, we feel that the current experimental set-up with a limited number of well-chosen samples and in-depth analysis of the obtained results allows drawing conclusions that are relevant for the practitioner. Another limitation of this study is that, considering the cultivation conditions used in this study, other relevant bacterial pathogens such as Histophilus somni and Mycoplasma bovis could not be isolated from the current clinical nBAL samples. However, H. somni is only rarely isolated because of poor viability and the fact that it is easily overgrown by other bacteria, either clinically relevant or not [3]. Although different studies are available describing the effect of storage conditions on the recovery of M. bovis, this was only investigated at low temperatures in milk samples [13, 14] or colostrum samples [15]. Therefore, further research into the effect of storage conditions on the recovery of M. bovis from nBAL samples is encouraged. Currently, the gold standard technique for identifying these fastidious bacteria is polymerase chain reaction (PCR). Since viability is not mandatory with this technique, it can be expected that the effect of storage conditions for identifying these pathogens will be less important in most veterinary labs using PCR to identify the latter pathogens. Moreover, maximizing the chance of isolating P. multocida and M. haemolytica is more critical since performing antimicrobial susceptibility testing in these species can be of major importance for appropriate antimicrobial treatment, while antimicrobial resistance is generally less prevalent in H. somni [16], although multiresistance has been described [17], or even not routinely tested for in M. bovis.
Conclusion
This study demonstrates that optimal M. haemolytica and P. multocida isolation rates from clinical nBAL samples are obtained after storage at 0 °C or 8 °C, provided that the sample is cultivated within 24 h after sampling. The maximum period a sample can be stored without an effect on the M. haemolytica and P. multocida isolation success varies and is dependent on the storage temperature and the degree of microbial contamination.
Methods
The sample size required to determine a 60% difference in isolation rate (80% vs 20%) with 80% power and 95% confidence for a 2-sided test was 8 positive nBAL samples per test group (Winepiscope 2.0, Zaragoza, Spain). Each test group comprises the presence of a clinically relevant bacterial pathogen, namely Pasteurella multocida and Mannheimia haemolytica. Therefore samples were taken until a minimum number of 8 positive culture results per test group was reached. An experimental study design was performed on 4 unrelated farms (3 beef, 1 dairy) between March and April 2018. Farms suffering from an acute outbreak of BRD were reported by local veterinarians and subsequently visited by the research staff. Animals to be sampled were selected based on previously described inclusion criteria [18]. Additionally, thoracic ultrasound examination was performed with a 7.5-MHz linear probe (Tringa Linear Vet, Esaote, the Netherlands) as previously described [19]. The definition for a case was the presence of a consolidated zone in the lung of ≥1 cm depth [20]. Animals that were treated with antimicrobials within 14 days prior to sampling were excluded from the study.
Cattle that met the inclusion criteria were sampled with the nBAL procedure as previously described [21]. Briefly, after disinfecting the nostril with 70% alcohol, a reusable home-made polytetrafluorethylene catheter adjusted with a 12-G catheter stylet was inserted in the nasal cavity and gently advanced, through larynx and trachea, into the bronchi. Next, 60 mL of sterile 0.9% NaCl was injected into the lungs and immediately aspirated (recovery of 30–50% of the fluid). Samples were transported at ambient temperature and further processed within 30 min after sampling.
Twenty mL of each nBAL sample was used for further analysis and was divided equally over four different 50 mL Falcon tubes after vortexing for 1 min (5 mL each). Each Falcon tube was incubated at a different temperature, all monitored with a thermometer, i.e. 0 °C +/− 1 °C (ice), 8 °C +/− 1 °C (refrigerator), 23 °C +/− 1 °C (room temperature) and 36 °C +/− 1 °C (incubator) for 0, 2, 4, 6, 8, 24 and 48 h. After each incubation period, the sample was vortexed for 30 s and 100 μL was transferred to an Eppendorf tube, already containing 900 μL phosphate buffered saline (PBS). Ten-fold dilutions were made of each sample for quantitative analysis as previously described [22]. From each dilution, 100 μl was inoculated on Columbia agar with 5% sheep blood (blood agar; Oxoid, Hampshire, UK) and incubated overnight at 35 °C +/− 2 °C in a 5% CO2 atmosphere. All macroscopically different colonies were counted and identified with MALDI-TOF MS as previously described [23]. A positive culture result was defined as the macroscopically visible presence of one or more clinically relevant Pasteurellaceae (P. multocida and M. haemolytica) colonies in pure, dominant or mixed cultures at T0 as previously described [3]. Contamination of these pure culture samples can occur over time due to incubation. Only samples with a positive culture at time point 0 h (T0) were included in the experiment. When no clinically relevant pathogen could be isolated at time points 2, 4, 6, 8, 24 or 48 h, the concentration of the pathogen isolated at T0 in this sample was set at 100 Colony forming units (CFU)/mL (being the detection limit of this plating procedure). A reduced viability of a certain bacterial pathogen was determined when the initial concentration declined linearly over time without an increase of the concentration of contaminants. An increased microbial contamination was determined as an increase of concentration of contaminants over time, until eventually the clinically relevant pathogen cannot be determined anymore.
The association between the different conditions for the isolation of Pasteurellaceae was determined by means of a multivariable logistic regression model with repeated measures (PROC GLIMMIX). Bonferroni corrections were used to compare between 4 groups. To determine the association between the presence of contaminants and isolation of M. haemolytica or P. multocida logistic regression was used (PROC LOGISTIC). Model validity was evaluated by the Hosmer-Lemeshow goodness-of-fit test for logistic models. Significance was set at P < 0.05. All data were collected in Microsoft Excel and statistical analysis was performed in SAS 9.4 (SAS Institute Inc., Cary, NY).
Availability of data and materials
The dataset used and analysed during the current study is available from the corresponding author on reasonable request.
Abbreviations
- BRD:
-
Bovine respiratory disease
- CFU:
-
Colony forming unit
- MALDI-TOF MS:
-
Matrix-assisted laser desorption/ionization-time-of- flight mass spectrometry
- nBAL:
-
Non-endoscopic bronchoalveolar lavage
- PCR:
-
Polymerase chain reaction
References
Snowder GD, Van Vleck LD, Cundiff LV, Bennett GL. Bovine respiratory disease in feedlot cattle: environmental, genetic and economic factors. J Anim Sci. 2006;84:1999–2008.
Pardon B, Catry B, Dewulf J, Persoons D, Hostens M, De Bleecker K, Deprez P. Prospective study on quantitative and qualitative antimicrobial and anti-inflammatory drug use in veal calves. J Antimicrob Chemother. 2012;67:1027–38.
Van Driessche L, Valgaeren B, Gille L, Boyen F, Ducatelle R, Haesebrouck F, Deprez P, Pardon B. A deep nasopharyngeal swab versus nonendoscopic bronchoalveolar lavage for isolation of bacterial pathogens from preweaned calves with respiratory disease. J Vet Intern Med. 2017. https://doi.org/10.1111/jvim.14668.
Smismans A, Verhaegen J, Schuermans A, Frans J. Evaluation of the Copan ESwab transport system for the detection of methicillin-resistant Staphylococcus aureus: a laboratory and clinical study. Diagn Microbiol Infect Dis. 2009;65:108–11.
Hansen MJ, Bertelsen MF, Dietz R, Sonne C, Bojesen AM. A simple and novel method for retrieval of Pasteurellaceae from swab samples collected in the field. Microbiol Open. 2013;2:795–7.
Rowe HA, Poxton IR, Donachie W. Survival of Mannheimia (Pasteurella) haemolytica in tracheobronchial washings of sheep and cattle. Vet Mic. 2001;81:305–14.
Tefera G, Smola J. Modification of Cary-Blair transport medium for Pasteurella multocida and Mannheimia haemolytica. Acta Vet BRNO. 2002;71:229–33.
van Rensburg E, du Preez JC, Kilian SG. Influence of the growth phase and culture medium on the survival of Mannheimia haemolytica during storage at different temperatures. J Appl Microbiol. 2004;96:154–61.
Tano E, Melhus Å. Evaluation of three swab transport systems for the maintenance of clinically important bacteria in simulated mono-and polymicrobial samples. APMIS. 2011;119:198–203.
Thomas AC, Bailey M, Lee MRF, Mead A, Morales-Aza B, Reynolds R, Vipond B, Finn A, Eisler MC. Insights into Pasteurellaceae carriage dynamics in the nasal passages of healthy beef calves. Sci Rep. 2019;9:11943.
Van Driessche L, Bokma J, Deprez P, Haesebrouck F, Boyen F and Pardon B. Rapid identification of respiratory bacterial pathogens from bronchoalveolar lavage fluid in cattle by MALDI-TOF MS. Sci Rep. 2019;9:18381.
Kugadas A, Poindexter J, Lee M-L, Bavananthasivam J, Call DR, Brayton KA, Srikumaran S. Growth of Mannheimia haemolytica: inhibitory agents and putative mechanism of inhibition. Vet Mic. 2014;174:155–62.
Al-Farha AA, Khazandi M, Hemmatzadeh F, Jozani R, Tearle R, Hoare A, Petrovski K. Evaluation of three cryoprotectants used with bovine milk affected with Mycoplasma bovis in different freezing conditions. BMC Res Notes. 2018;11:216.
Boonyayatra S, Fox LK, Besser TE, Sawant A, Gay JM. Effects of storage methods on the recovery of Mycoplasma species from milk samples. Vet Mic. 2010;144:210–3.
Gille L, Boyen F, Van Driessche L, Valgaeren B, Haesebrouck F, Deprez P, Pardon B. Effect of freezer storage time and thawing method on the recovery of Mycoplasma bovis from bovine colostrum. J Dairy Sci. 2017;101:609–13.
DeDonder KD, Apley MD. A literature review of antimicrobial resistance in pathogens associated with bovine respiratory disease. Anim Health Res Rev. 2015;16:125–34.
Bhatt K, Timsit E, Rawlyk N, Potter A, Liljebjelke K. Integrative conjugative element ICEHs1 encodes for antimicrobial resistance and metal tolerance in Histophilus somni. Front Vet Sci. 2018;5:153.
Pardon B, Alliet J, Boone R, Roelandt S, Valgaeren B, Deprez P. Prediction of respiratory disease and diarrhea in veal calves based on immunoglobulin levels and the serostatus for respiratory pathogens measured at arrival. Prev Vet Med. 2015;120:169–76.
Ollivett TL, Caswell JL, Nydam DV, Duffield T, Lesli KE, Hewson J, Kelton D. Thoracic ultrasonography and bronchoalveolar lavage fluid analysis in Holstein calves with subclinical lung lesions. J Vet Int Med. 2015;29:1728–34.
Buczinski S, Forte G, Belanger A. Short communication: ultrasonographic assessment of the thorax as a fast technique to assess pulmonary lesions in dairy calves with bovine respiratory disease. J Dairy Sci. 2013;96:4523–8.
Van Driessche L, Valgaeren B, De Schutter P, Gille L, Boyen F, Ducatelle R, Deprez P, Pardon B. Effect of sedation on the intrapulmonary position of a bronchoalveolar lavage catheter in calves. Vet Rec. 2016;179:18.
Van Hecke LL, Hermans K, Haspeslagh M, Chiers K, Pint E, Boyen F, Martens AM. A quantitative swab is a good non-invasive alternative to a quantitative biopsy for quantifying bacterial load in wounds healing by second intention in horses. Vet J. 2017;225:63–8.
Kuhnert P, Bisgaard M, Korczak BM, Schwendener S, Christensen H, Frey J. Identification of animal Pasteurellaceae by MALDI-TOF mass spectrometry. J Microbiol Meth. 2012;89:1–7.
Acknowledgements
The authors would like to acknowledge the farmers who participated in this field study.
Funding
This study was funded by a PhD Fellowship of the Research Foundation-Flanders (FWO-1S52616N). The MALDI-TOF mass spectrometer was financed by the Research Foundation Flanders (FWO-Vlaanderen) as Hercules project G0H2516N (AUGE/15/05). The funders had no role in the design of the study, the collection, analysis, and interpretation of data nor in writing the manuscript.
Author information
Authors and Affiliations
Contributions
LVD designed and conducted all experiments, collected literature data, analysed data, prepared figures and prepared the paper. CDN conducted all experiments, collected literature data, analysed data and prepared figures. FB designed the study, analysed data and supervised the work. BP designed the study, analysed data, prepared figures and supervised the work. KvL collected literature data. FH analysed data. The author(s) read, reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All procedures were approved by the ethical committee of the Faculty of Veterinary Medicine, Ghent University, Belgium (EC 2016/20). Verbal informed consent was obtained from all participants by telephone (veterinary officers and farmers), which was approved by the ethical committee of the Faculty of Veterinary Medicine, Ghent University, Belgium (EC 2016/20).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Van Driessche, L., De Neve, C., Haesebrouck, F. et al. Storage time and temperature affect the isolation rate of Mannheimia haemolytica and Pasteurella multocida from bovine bronchoalveolar lavage samples. BMC Vet Res 16, 238 (2020). https://doi.org/10.1186/s12917-020-02456-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-020-02456-7