Abstract
Background
Multi-drug methicillin-resistant Staphylococcus pseudintermedius (MRSP) detection is rapidly increasing in microbial specimens from pets across Europe. MRSP has also been isolated from bitches and newborns in dog breeding kennels. This study assessed whether MRSP lineage differs between breeding kennels and is maintained over time. Post-partum bitches (at day 3 vaginal and day 3, 9 and 35 milk samples) and their litters (at day 3, 9 and 35 oral and abdominal skin samples) from two Italian breeding kennels (A and B) were sampled and MRSP was subsequently characterized via whole-genome sequencing and antibiotic susceptibility testing. The study was carried out from October 2014 to March 2016 and included successive parturitions from the same animals.
Results
The analysis revealed different situations in both investigated kennels. In kennel A, circulating strains were from 7-locus sequence types ST688, ST258 and closely related isolates of ST71, which included most isolates. In kennel B, only a new isolate, ST772, was detected. In addition, most isolates from both kennels had multi-resistant antibiotic profiles. MRSP was only isolated from litters of MRSP-positive bitches, thus suggesting that bitch-litter transmission is likely.
Conclusions
Our data show that MRSP circulation can differ in different settings, that several clonal lineages can circulate together, and that vertical transmission appears common. MRSP colonization did not affect the health conditions of the bitches or of their litters.
Similar content being viewed by others
Background
Staphylococcus pseudintermedius belongs to the bacterial skin flora of canines and is a common opportunistic pathogen of dogs [1]. In addition, S. pseudintermedius is commonly detected in the genital organs of both healthy dogs and dogs with reproductive problems, including in the vaginas of bitches in the pre- and postpartum periods [2, 3]. It has been linked to neonatal mortality due to necrotising dermatitis, pododermatitis, acute suppurative interstitial pneumonia [4], toxaemia and septicaemia [5].
Due to the importance of S. pseudintermedius as an opportunistic pathogen, it is worrisome that multi-drug methicillin-resistant S. pseudintermedius (MRSP) detection has rapidly increased in microbiological specimens from pets across Europe in recent years [6, 7]. MRSP is often regarded as a threat due to limited therapeutic options and because it frequently shows resistance profiles to three or more antimicrobial classes [8, 9]. Among European isolates, the multi-resistant clonal lineage ST71-J-t02-II–III is dominant [10], but other emerging clones, such as ST258, have also been detected in some European countries [11,12,13]. MRSP has also occasionally been isolated from the colostrum and milk of breeding bitches as well as from puppies [5, 14]. However, data are lacking on the relatedness and transmission of MRSP lineages isolated from bitches and their offspring in breeding kennels. Therefore, this study assessed whether the same MRSP lineage is present in bitches and their litters in different breeding kennels and is maintained over time.
Methods
Animals and sampling
Two breeding kennels (A and B) in Piedmont (Italy) were identified as MRSP-positive in a study investigating the microbial flora of bitches and their litters, conducted from October 2014 to March 2016. The current investigation characterized and compared MRSP isolates identified in that study from bitches and their offspring (Table 1).
The sampling scheme for the bitches was as follows: vaginal swabs were collected on postpartum day 3, and milk swabs were collected from all mammary glands on postpartum days 3, 9 and 35. The puppies were sampled at the same frequency at two sites: the abdominal skin cranial to the umbilicus (rubbing the swab for 5 s on a 4 cm2 area) and the mouth (rubbing the swab on the oral mucosa). All swabs (Copan Italia, Brescia, Italy) were placed in Amies medium (Copan Italia, Brescia, Italy), transported to the laboratory at environmental temperature and cultured within 48 h.
The study was performed in accordance with the guidelines for the care and use of animals of the Department of Veterinary Science of the University of Turin. Informed consent was obtained from the dog breeders prior to testing.
MRSP identification
The swabs were directly plated onto 5% sheep blood agar (SBA, Oxoid, Basingstoke, UK), for a general assessment of bacterial growth, especially that of staphylococci, and in 6.5% NaCl nutrient broth (Mueller Hinton broth T073–BIOKAR Diagnostic, Allonne, France) and incubated overnight at 37 ± 1 °C. After incubation, 10 μl of the salt broth was inoculated into a selective medium (CHROMagar™ MRSA II plates, Heidelberg, Germany). Staphylococci grown on blood agar plates were presumptively identified based on colony morphology, Gram staining, catalase tests, haemolysis and pigment production. Coagulase-positive staphylococci were identified by tube coagulase testing on rabbit plasma (Istituto Zooprofilattico delle Venezie, Legnaro, Italy) and incubated at 35 °C ± 1 °C for 4 h. One to three isolated colonies were added to 0.5 ml of rabbit plasma in a tube and incubated at 35 °C. Those samples testing negative after 4 h were maintained at room temperature and read again after 18–24 h.
All colonies with pink, light pink or white colouring after 24–48 h on CHROMagar™ MRSA II plates were regarded as suspect methicillin-resistant coagulase-positive staphylococci. Five colonies grown on selective medium plates after enrichment were selected for confirmation of the presence of the mecA gene by conventional PCR [15]; staphylococci species identification was also performed by PCR [16].
Antimicrobial susceptibility tests
Minimum inhibitory concentration (MIC) was determined using broth microdilution (Sensititre COMPAGN1F, Thermo Fisher Scientific, Waltham, MA USA). Isolates were tested for susceptibility to oxacillin and a panel of 18 antimicrobials belonging to nine antimicrobial classes: beta-lactams (penicillin, ampicillin, amoxicillin/clavulanic acid, ticarcillin, ticarcillin/clavulanic acid, cefazolin, cefpodoxime, ceftiofur), tetracyclines (doxycycline), diaminopyrimidine-sulphonamide combinations (trimethoprim/sulfathiazole), aminoglycosides (gentamicin, amikacin), macrolides (erythromycin), lincosamides (clindamycin), fluoroquinolones (enrofloxacin, marbofloxacin), amphenicols (chloramphenicol) and rifamycins (rifampicin).
Antimicrobial susceptibility results were interpreted per the Clinical Laboratory Standards Institute [17, 18]. In case of no clinical breakpoints set for S. pseudintermedius in dogs, we used breakpoints for Staphylococcus spp. in dogs and, when unavailable in dogs, we used other species (horse and cattle) or human breakpoints (Table 2). For ticarcillin and ticarcillin/clavulanic acid, the [19] were used, and S. aureus DSMZ 11729 was used as a quality control.
Genotypic characterization using whole genome sequencing
Whole genome sequencing (WGS) was carried out on only fifteen isolates due to economic limitations. The isolates were selected to represent MRSP-positive bitch-litter units from different sampling times and successive parturitions of the same bitch (Table 1). A sixteenth isolate dating from 2013, cultured from the milk of a bitch who was again MRSP-positive in 2014–2016, was also included in the WGS analysis [[14]; bitch N° 3].
DNA was extracted from agar-plated colonies using an EZ1 Advanced and EZ1 DNA tissue kit (Qiagen, Hilden, Germany), and DNA concentration was determined using a Qubit HS DNA kit (Life Technologies, Carlsbad, CA, USA). Sequencing libraries were created using the Nextera XT kit (Illumina, San Diego, CA, USA), and 250-bp paired-end sequencing was performed with a MiSeq sequencer (Illumina). Genome assembly was performed with SPAdes v.3.9.1 using the ‘—careful’ parameter [20] and an average coverage of 200×. Whole-genome sequence reads for the 16 isolates were deposited in the European Nucleotide Archive with the study accession number PRJEB19741. The 7-loci sequence types were determined by uploading the assemblies to the pubMLST server [21].
Core genome MLST (cgMLST) was performed using SeqSphere+ v3.5.0 software (Ridom GmbH, Münster, Germany), with cgMLST target genes extracted from the MRSP E140 genome (NCBI Accession: NZ_ANOI). The resulting cgMLST scheme consisted of 2183 genes for phylogenetic analysis (Additional file 1: Table S1). For comparison, unrelated MRSP genomes were added to the analysis: one Danish ST71 (E140), one German ST71 (E104, NCBI accession: LAWU), five genomes representing different ST71-clusters from a Swedish study [22] and five reference isolates, 081661 (NCBI accession: NZ_CP016073), 9,841,787 (NCBI accession: JTKQ), 9,841,998 (NCBI accession: JTKP), 2001–08-299 (NCBI accession: JTKO) and NA45 (NCBI accession: NZ_CP016073). To visualize the number of allelic differences between the genomes, minimum spanning trees were created in SeqSphere+.
Using task templates based on the S. aureus microarray (Alere Technologies GmbH, Jena, Germany) available in SeqSphere+, occurrence of genes involved in regulation, resistance and virulence was assessed. Occurrence of genes encoding antibiotic resistance was also verified using the Comprehensive Antibiotic Resistance Database (https://card.mcmaster.ca/, 2017-07-03). In addition, occurrence of the S. pseudintermedius toxin genes lukI/S (X79188), siet (AB099710), SE-INT (AB116378), secCanine, (U91526), expA (AB489850) and expB (AB569087) was determined by read mapping using Bowtie2 [23].
Results
From October 2014 to March 2016, MRSP was isolated from two of five bitches tested in kennel A and from four of eleven bitches tested in kennel B, as well as from their puppies. All the bitches were healthy and no puppy died or showed pathological signs during observation.
In kennel A, bitches A1 and A2 were positive for MRSP in either vaginal or milk swabs. A1 was positive in two successive parturitions, 1 year apart, and had already had MRSP-positive milk after a previous parturition in 2013 [[14]; bitch N° 3]. A1 had eleven puppies in 2014 and ten in 2015/16, and of these, four and six were tested respectively, with all being positive for MRSP at least once in the oral mucosa or abdominal skin (Table 1). A2 had one pup in 2014 and two in 2015/16, and all three were tested and shown to be MRSP-positive at least once (Table 1).
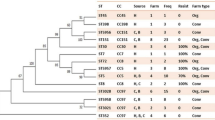
All S. pseudintermedius isolates from kennel A were MRSP, for a total of 22. Of these, ten were selected for WGS. With two exceptions, every isolate belonged to the seven-gene multi-locus sequence type (7-MLST) ST71. One isolate from bitch A1 on day 35, after the last parturition, (Table 1, isolate 9778) belonged to the new type ST688, which shares five of seven alleles with ST71, while the isolate from the same bitch in 2013 (Table 1, isolate 9771) belonged to type ST258. ST258 is distinct from both ST71 and ST688 based on the 7-MLST scheme and the cgMLST (Fig. 1). Based on cgMLST, all ST71 isolates were genetically related, differing only in up to four alleles of the 2100 genes when compared pairwise (data not shown). Isolates from kennel A formed a separate cluster from other MRSP-ST71 genomes available from public databases (Fig. 1).
Minimum spanning tree of the core genome MLST results from all 16 MRSP isolates (green for kennel A and yellow for kennel B) and from five Swedish isolates representing five different clusters of ST71 isolates (blue) and reference isolates from GenBank (white). The white isolates comprise strains (with NCBI accessions in parentheses): REF 1: 081661 (NZ_CP016073), REF 2: E104 (LAWU), REF 3: E140 (ANOI), REF 4: 9841787 (JTKQ), REF 5: 9841998 (JTKP), REF 6: 2001–08-299 (JTKO), REF 7: NA45 (NZ_CP016073). The results are based on analysis of 1626 genes. The numbers represent the number of allele differences between isolates, and the lines are not proportional to the number
In kennel B, bitches B1-B4 were MRSP-positive in either vaginal or milk swabs. Two puppies from B1 and B2, from two litters of two, yielded MRSP-positive oral mucosa swabs on day 3 (Table 1). MRSP-positive bitches and puppies also carried non-methicillin-resistant strains.
Of the eight MRSP isolates identified, five were selected for WGS, following the same criteria as for kennel A. The five sequenced isolates all belonged to the new type ST772. Based on cgMLST, these isolates clearly differed from those in kennel A but were less diverse, differing only by 1–7 alleles (see Fig. 1).
Most isolates genotyped from kennel A were resistant to seven antimicrobial classes other than beta-lactams, with susceptibility only to rifampicin (Table 3). Reduced susceptibility to amikacin was shown by six out of nine ST71 isolates (Table 3). The phenotypic resistance profile was confirmed by the presence of resistance genes. The ST71 isolates all carried the methicillin-resistance gene mecA, the beta-lactamase gene blaZ, and genes encoding resistance to aminoglycosides aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa and aad [6], macrolide erm(B), streptothricin sat-4 and trimethoprim dfrG. All isolates were positive for tetK, which encodes for tetracycline resistance, except isolate 97711 from the oral mucosa of a puppy from bitch A2 at 35 days after the second parturition. Based on cgMLST, the 97711 isolate differed by only one allele from isolate 97710, which was isolated simultaneously from bitch A2. The lack of tetK was due to the loss of a plasmid virtually identical to the 4.5-kb plasmid pT181 (J01764.1), first described in S. aureus [24]. Isolate 97711 was indeed susceptible to doxycycline in vitro. The ST688 isolate, that was susceptible to gentamycin and amikacin, lacked the genes aac(6′)-Ie-aph(2″)-Ia and tetK but carried tetM for tetracycline resistance and cat for chloramphenicol resistance (Table 3). ST258, dating from 2013, was sensitive to several tested antimicrobials. It showed a similar resistance gene profile to ST688 but lacked cat and carried qacC, which mediates resistance to antiseptics, disinfectants and beta-lactam antibiotics [25].
Similar to the kennel A isolates, all kennel B isolates showed multi-resistance profiles, although they differed in fluoroquinolone susceptibility. Most of them were resistant to both amikacin and rifampicin (Table 4). They were all identical in the genes encoding antibiotic resistance, namely, aac(6′)-Ie-aph(2″)-Ia, aph(3′)-IIIa, aad [6], dfrG, sat-4 and ermB.
All MRSP isolates from kennels A and B contained the toxin genes lukI/S, siet and SE-INT.
Discussion
WGS revealed two different situations in the investigated kennels. In kennel A, multiple strains continued to circulate, as previously evidenced by PFGE in 2013 [14], while in kennel B, the isolates were highly related, with a new ST772 type circulating. A possible explanation for the difference between the two kennels could be that the introduction and spread of MRSP in kennel B was more recent or that the selection pressure was weaker, as suggested by the concomitant isolation of MSSP and MRSP from the bitches and puppies of kennel B. Both kennels represent two distinct settings with different factors, among which could be the amount and type of antimicrobial use, which may have influenced the selection and spread of the different MRSP strains. Although beta-lactams are the first-choice antimicrobials for treating breeding bitches in both investigated kennels, we have no precise data on the general frequency/dose/reasons that these agents were administered.
MRSP isolates are commonly multi-drug resistant, but resistance to amikacin and rifampicin is an emerging trait. Amikacin-resistant strains have been isolated with increasing frequency from canine patients [26] but not from healthy dogs. In S. pseudintermedius, the most common amikacin resistance gene was found to be aph(3′)-IIIa [26], which is present in the isolates detected here. Aminoglycosides such as amikacin are among the last choices of treatment for staphylococcal infections and have not been used on the bitches included in this study. In addition, rifampicin resistance, due to mutation in the rpoB gene, is reported in MRSP isolated from infected dogs treated with rifampicin [27], which is not the case with the healthy breeding bitches in this study.
The detection of the gene qacC in the isolate dating from 2013 is also remarkable. This plasmid-borne gene, responsible for transporting quaternary ammonium compounds and ethidium bromide out of the cell, has been detected in S. aureus and coagulase-negative staphylococci but not in S. pseudintermedius [28]. Quaternary ammonium disinfectants are commonly used in kennels, and repeated sub-lethal exposure could have selected for the expression of this efflux pump. However, a gene-transfer from other Staphylococcus species could also have been a possible explanation for the presence of qacC [28].
Dogs with clinical MRSP infections can be carriers for several months after acquisition [29, 30], but data on MRSP colonization persistence and maternal-offspring transmission are scant. Previous studies have reported that individual dogs can be either persistent or intermittent carriers of highly genetically diverse MRSP strains, isolated from different body sites [1]. We therefore cannot exclude the possibility of the simultaneous presence of different STs in the bitch that carried ST258 in 2013 [14] and ST688 in 2014. Moreover, ST71 was isolated from her puppies; although we could hypothesize an environmental origin for ST71, the maternal source is also possible. The chosen sampling locations may have influenced these results, and testing different or additional body sites could have yielded more MRSP genotypes on a single animal at the same sampling time [30].
In our work, MRSP was only sporadically isolated from mothers and puppies during the 35-day sampling period. Furthermore, MRSP was not always present simultaneously in the bitches and their puppies. Interestingly, however, we showed that MRSP occurrence in the puppies appeared to be connected to MRSP-positive bitches. Apart from two exceptions in kennel B, MRSP was isolated at different times from all litters of the colonized bitches and from most of the tested puppies from the colonized litters. A potential transmission from bitch to offspring, or at least a spread within the kennel, appears to be likely due to the high genetic relatedness of the strains within a kennel. Previous studies have shown that puppies are colonized with S. pseudintermedius almost immediately after birth and that the first contact is from the buccal flora of their mother because the amniotic membrane is generally intact when puppies are born [2, 31]. Furthermore, close contact during nursing contributes to the rise of cutaneous S. pseudintermedius flora in puppies [31]. Another likely source for the colonization of puppies is the milk and the mammary glands. S. pseudintermedius is commonly isolated from the milk of healthy post-partum bitches due to the sinus system of the gland having been colonized [32, 33]. In the current study we cannot exclude the milk having been contaminated by the skin during sample collection because samples were milked cumulatively from every mammary gland.
Conclusions
Our data show that MRSP circulation can differ in different settings, from the simultaneous detection of multiple strains to the isolation of a single genotype, such as with the case of the new ST772 type.
Worth noticing is the detection of amikacin- and rifampicin-resistant isolates carried by healthy bitches and puppies that had not been treated with these antimicrobials.
Vertical bitch-litter transmission appears common since MRSP was isolated at different sampling times from most of the litters of the colonized bitches and from most puppies tested from the colonized litters. It is also likely that the bitches were the primary source since the MRSP from the puppies was only isolated from litters of MRSP-positive bitches.
Our study only covered the period from birth to weaning; thus, it would be interesting to assess how long the MRSP is carried, whether the bitches are colonized in different locations during and after the end of lactation, and how different locations may affect potential transmission to the puppies.
References
Bannoehr J, Guardabassi L. Staphylococcus pseudintermedius in the dog: taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet Dermatol. 2012;23:253–66.
Allaker RP, Jensen L, Lloyd DH, Lamport AI. Colonization of neonatal puppies by staphylococci. Brit Vet J. 1992;148:523–8.
Bjurström L, Linde-Forsberg C. Long-term study of aerobic bacteria of the genital tract of breeding bitches. Am J Vet Res. 1992;53:665–9.
Philbey AW, Taylor DJ, Robb A, Gibbons JF, Irvine RM, Thompson H. Staphylococcal dermatitis/pododermatitis and septicaemia in neonatal puppies. Vet Rec. 2013;173:424–6.
Rota A, Milani C, Drigo I, Drigo M, Corrò M. Isolation of methicillin-resistant Staphylococcus pseudintermedius from breeding dogs. Theriogenology. 2011;75:115–21.
Ruscher C, Lübke-Becker A, Semmler T, Wleklinski CG, Paasch A, Soba A, Stamm I, Kopp P, Wieler LH, Walther B. Widespread rapid emergence of a distinct methicillin- and multidrug-resistant Staphylococcus pseudintermedius (MRSP) genetic lineage in Europe. Vet Microbiol. 2010;144:340–6.
Pires Dos Santos T, Damborg P, Moodley A, Guardabassi L. Systematic review on global epidemiology of methicillin-resistant Staphylococcus pseudintermedius: inference of population structure from multilocus sequence typing data. Front Microbiol. 2016;7:1599.
Beever L, Bond R, Graham PA, Jackson B, Lloyd DH, Loeffler A. Increasing antimicrobial resistance in clinical isolates of Staphylococcus intermedius group bacteria and emergence of MRSP in the UK. Vet Rec. 2015;176:172.
Borjesson S, Landen A, Bergstrom M, Andersson UG. Methicillin-resistant Staphylococcus pseudintermedius in Sweden. Microb Drug Resist. 2012;18:597–603.
Perreten V, Kadlec K, Schwarz S, Grönlund Andersson U, Finn M, Greko C, Moodley A, Kania SA, Frank LA, Bemis DA, Franco A, Iurescia M, Battisti A, Duim B, Wagenaar JA, van Duijkeren E, Weese JS, Fitzgerald JR, Rossano A, Guardabassi L. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J Antimicrob Chemother. 2010;65:1145–54.
Osland AM, Vestby LK, Fanuelsen H, Slettemeås JS, Sunde M. Clonal diversity and biofilm-forming ability of methicillin-resistant Staphylococcus pseudintermedius. J Antimicrob Chemother. 2012;67:841–8.
Haenni M, de Moraesa NA, Alves N, Châtrea P, Médailleb C, Moodleyc A, Madeca JY. Characterisation of clinical canine meticillin-resistant and meticillin-susceptible Staphylococcus pseudintermedius in France. J Global Antimicrob Resistance. 2014;2:119–23.
Duim B, Verstappen KM, Broens EM, Laarhoven LM, van Duijkeren E, Hordijk J, de Heus P, Spaninks M, Timmerman AJ, Wagenaar JA. Changes in the population of methicillin-resistant Staphylococcus pseudintermedius and dissemination of antimicrobial-resistant phenotypes in the Netherlands. J Clin Microbiol. 2016;54:283–8.
Rota A, Corrò M, Drigo I, Bortolami A, Börjesson S. Isolation of coagulase-positive staphylococci from bitches’ colostrum and milk and genetic typing of methicillin-resistant Staphylococcus pseudintermedius strains. BMC Vet Res. 2015;11:160.
Louie L, Goodfellow J, Mathieu P, Glatt A, Louie M, Simor AE. Rapid detection of methicillin-reistant staphylococci from blood culture bottles by using a multiplex PCR assay. J Clin Microbiol. 2002;40:2786–90.
Bannoehr J, Franco A, Iurescia M, Battisti A, Ross Fitzgerald J. Molecular diagnostic identification of Staphylococcus pseudintermedius. J Clin Microbiol. 2009;47:469–71.
CLSI VET01-S3. Performance Standards For Antimicrobial Disk and Dilution Susceptibility Tests For Bacteria Isolated From Animals. Third Informational Supplement. Wayne: Clinical and Laboratory Standards Institute; 2015.
CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria isolated From Animals: Approved Standard–Fourth Edition. CLSI VET01-A4. Wayne (PA): The Committee; 2013. CLSI document VET01-S2, 2013.
European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2018: http://www.eucast.org/. Accessed 2 Jan 2018.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77.
Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595.
Windahl U, Ågren J, Holst BS, Börjesson S. Colonization with methicillin-resistant Staphylococcus pseudintermedius in multi-dog households: a longitudinal study using whole genome sequencing. Vet Microbiol. 2016;30:8–14.
Langmead B, Salzberg S. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–9.
Khan SA, Novick RP. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid. 1983;10:251–9.
McDonnell G, Russell AD. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev. 1999;12:147–79.
Gold RM, Cohen ND, Lawhon SD. Amikacin resistance in Staphylococcus pseudintermedius isolated from dogs. J Clin Microbiol. 2014;52:3641–6.
Kadlec K, van Duijkeren E, Wagenaar JA, Schwarz S. Molecular basis of rifampicin resistance in methicillin-resistant Staphylococcus pseudintermedius isolates from dogs. J Antimicrob Chemother. 2011;66:1236–42.
Wassenaar TM, Ussery D, Nielsen LN, Ingmer H. Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. Eur J Microbiol Immunol (Bp). 2015;5:44–61.
Laarhoven LM, de Heus P, van Luijn J, Duim B, Wagenaar JA, van Duijkeren E. Longitudinal study on methicillin-resistant Staphylococcus pseudintermedius in households. PLoS One. 2011;6:e27788.
Windahl U, Reimegard E, Holst BS, Egenvall A, Fernstrom L, Fredriksson M, Trowald-Wigh G, Andersson UG. Carriage of methicillin-resistant Staphylococcus pseudintermedius in dogs-a longitudinal study. BMC Vet Res. 2012;8:34.
Saijonmaa-Koulumies LE, Myllys V, Lloyd DH, Saijonmaa-Koulumies LE, Lloyd DH. Diversity and stability of the Staphylococcus intermedius flora in three bitches and their puppies. Epidemiol Infect. 2003;131:931–7.
Kuhn G, Pohl S, Hingst V. Investigations on bacteria in the milk of healthy lactiferous bitches [in German]. Berl Munch Tierarztl Wochenschr. 1991;104:130–3.
Schäfer-Somi S, Spergser J, Breitenfellner J, Aurich JE. Bacteriological status of canine milk and septicaemia in neonatal puppies-a retrospective study. J Vet Med B Infect Dis Vet Public Health. 2003;50:343–6.
Funding
The work was funded by the Italian Ministero dell’Istruzione, Università e Ricerca (ex 60% 2016).
Availability of data and materials
Whole-genome sequence reads for the 16 isolates were deposited in the European Nucleotide Archive with the study accession number PRJEB19741.
Author information
Authors and Affiliations
Contributions
This study was set up by all the authors. AR conducted the clinical portion, while MC, JS and SB performed the laboratory portion; MC isolated the bacteria, and JS and SB performed the genotyping. Project funding was acquired by AR. AR drafted the manuscript, which was critically revised by all authors who have also read and approved the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study analysed samples collected in a previous investigation that had been performed in accordance with the guidelines for the care and use of animals of the Department of Veterinary Science of the University of Turin. The use of the samples followed an informed consent that was obtained from the dog owners.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Table S1. (XLS 260 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Corrò, M., Skarin, J., Börjesson, S. et al. Occurrence and characterization of methicillin-resistant Staphylococcus pseudintermedius in successive parturitions of bitches and their puppies in two kennels in Italy. BMC Vet Res 14, 308 (2018). https://doi.org/10.1186/s12917-018-1612-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-018-1612-z