Abstract
Background
Flavobacterium psychrophilum causes serious fish diseases such RTFS and BCWD, affecting the aquaculture industry worldwide. Commercial vaccines are not available and control of the disease depends on the use of antibiotics. Reliable methods for detection and identification of different isolates of this bacterium could play an important role in the development of good management strategies. The aim of this study was to identify genetic markers for discrimination between isolates. A selection of eight VNTRs from 53 F. psychrophilum isolates from Norway, Chile, Denmark and Scotland were analyzed. The results were compared with previous work on the same pathogen using MLST for genetic differentiation.
Results
The VNTR analysis gave a separation between the F. psychrophilum isolates supporting the results of previous MLST work. A higher diversity was found among the Chilean isolates compared to those from Norway, which suggests a more homogenous reservoir in Norway. Transgenerational transmission of F. psychrophilum from other countries, exporting salmon embryos to Chile, may explain the differences in diversity. The same transmission mechanisms could also explain the wide geographical distribution of identical isolates in Norway. But, this could also be a result of movement of smolts and embryos. The selected VNTRs are stable genetic markers and no variation was observed after several passages on agar plates at different temperatures.
Conclusions
These VNTRs are important additions for genotyping of F. psychrophilum isolates. Future studies on VNTRs of F. psychrophilum should include isolates from more host species from a wider geographical area. To get a more robust genotyping the VNTRs should be used in concert with MLST. Future studies of isolates with high and low virulence should focus on identifying virulence markers using VTNRs and MLST.
Similar content being viewed by others
Background
Flavobacterium psychrophilum causes bacterial cold water disease (BCWD) and rainbow trout fry syndrome (RTFS) in the fresh water phase of salmonid production worldwide [1]. Although some commercial vaccines are available (J. Battaglia com. pers.) flavobacteriosis is still a major problem in the salmonid production in Chile, and the control of the disease is dependent on the use of antibiotics [2]. Another approach is the use bacteriophages that have been tested with promising effects and use of probiotics and modified fish diets represent other options [3, 4].
Since the first reports of flavobacteriosis, the disease caused by F. psychrophilum (former name Cytophaga psychrophila) in the 1940s in North America, the pathogen has emerged in several countries causing severe outbreaks on salmonids and other fresh water species [5, 6]. It has been shown that F. psychrophilum can be transgenerational transmitted and, to prevent spreading and disease outbreaks it is necessary to develop specific and sensitive methods for detection of the bacterium [7].
Differences in virulence among F. psychrophilum isolates will have implications for management strategies and vaccine development [8]. These management strategies are dependent on the correct identification of the virulence and the source of origin the isolates. Based on existing knowledge the best approach for developing such identification tools will to involve knowledge about the genetics of the bacteria. Several methods have been developed for genetic differentiation of other fish pathogenic bacteria [9, 10]. The most commonly used methods are restriction fragment length polymorphism (RFLP), random amplified polymorphism (RAPD), 16S rDNA sequencing, housekeeping genes sequencing, MLST and VNTRs [11–17]. These methods or a selection of them can also be used for identification of host specificity and studies of geographical distribution, molecular epizootiology, and antibiotic resistance.
Use of variable number of tandem repeats (VNTR) are among the most discriminating and least time consuming of the genotyping methods. This method has been widely used during the last 15 years for genotyping of bacteria causing diseases in humans, domestic animals and vegetables [18–22]. In aquaculture, the method has been used for genotyping of Atlantic salmon Salmo salar [23], and the parasitic copepod Caligus rogercresseyi Boxshall and Bravo 2000 [24], but also for genotyping of other fish pathogenic bacteria [14, 25–27].
The aim for this study was to identify VNTRs that can be used to generate a genotyping system for separation of F. psychrophilum isolates. This VNTR system was applied to a set of isolates from Norway and Chile.
Results
Variable number of tandem repeats (VNTR)
The 12 pairs of VNTR-primers were tested using DNA extracted from the type strain (NCIMB 1947), five Norwegian, and five Chilean isolates of F. psychrophilum. Among this initial group of 11 isolates, only eight of the 12 amplified VNTRs loci showed variation. The remaining four (VNTR-2, VNTR-3, VNTR-4 and VNTR-12), were not tested for the other isolates. For some isolates, the primers for VNTR-7 and VNTR-13 did not produce the expected PCR products, but a new set of primers yielded these products with the exception of the isolate No10-35-T-W. The accession numbers of all the obtained VNTRs are included in the Additional file 1.
The eight VNTR loci gave variation from one to nine repetitions among the isolates (n = 53) included a table with all the allelic VNTR profiles for all the isolates is presented in the Additional file 2. The type strain and some isolates from Norway and Chile present two types of deletions in VNTR-7, where the complete repetitive sequence is TTAAAAA. One variant is TAAAA and the other is AAAA with no repetitions. The VNTRs, related to the genome of JIP02 /86, show that they are located in both intergenic and intragenic positions. In the case of VNTR-10, the repeats are located in the gene associated with gliding motility and encode for the GldL protein (Table 3). No variation in the number of repeats and nucleotide positions in the eight VNTRs loci studied were observed when the VNTR stability of isolate No12-49-As-Op was tested after 12 passages on agar plates at two different temperatures (4 °C and 15 °C).
The obtained sequencing data for the VNTRs were used to prepare profiles for the isolates of F. psychrophilum. These profiles revealed 25 different VNTR sets (VS) (Supplementary material). This VNTR polymorphism constituted the basis for the establishment of a multiple-loci VNTR analysis (MLVA) system for separation of the isolates.
Grouping of the F. psychrophilum isolates
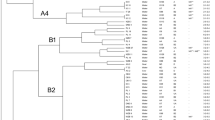
The relationships between the F. psychrophilum isolates, based on VNTR analysis, are presented in Fig. 1. The majority of the isolates from Norway and Chile group in separated clades. However, eleven European isolates are found among the Chilean clades (A, B, and C), while the majority of the Norwegian isolates group in distinct clades with the type species. A second group of the Chilean isolates constitute the most basal clade (F). These results are also to a large extent supported by the PCA, Fig. 2.
The genetic relationships of the isolates included in this study. Relationship among 53 isolates of F. psychrophilum based on allelic differences at eight VNTR loci. This unrooted ultrameric NJ tree show know topography between Norwegian and Chilean isolates. Codes of are explained in the table included in Additional file 3
PCA plot of all 53 isolates of F. psychrophilum, from Norway (red), Chile (blue), Denmark (green) and Scotland (violet), type strain NCIMB 1947 and of F. psychrophilum JIP02/86. The figure also includes the different VNTRs analysed. The isolate No12-49-As-Op tested for VNTR stability (4 and 15 °C) showed the same VT profiles in both temperatures Additional file 2, therefore they are not shown in this figure
Among the 53 isolates included in the study only one group (clade D) were obtained from one host species (Salmo salar) only. Another clade (C) was dominated by isolates from rainbow trout and a third clade (B) by isolates from Atlantic salmon. Both these clades consisted of a majority of Chilean isolates. Clade E consists of isolates obtained from wild trout (Salmo trutta) and Atlantic salmon from river Eira, western of Norway, and the type species (NCIMB 1947) from coho salmon. The PCA confirm this distribution.
Isolates of F. psychrophilum were obtained from both healthy fish and fish showing signs of flavobacteriosis. Based on VNTR analysis none of the clades show clear tissue specificity. However, both analyses identified one group of possible pathogenic Norwegian isolates obtained from Atlantic salmon suffering flavobacteriosis (clade D, VS8, in Figs. 1 and 2). Possible pathogenic isolates from moribund fish collected in four different countries are also present in clade C. Clade E consists of isolates from both healthy and moribund wild salmonids.
Discussion
Several techniques, based on nucleotide sequencing targeting different genes, have been employed in attempts to separate isolates of F. psychrophilum, and with highly variable results [28–30]. Among these, multi locus sequence typing (MLST) has been among the most promising methods [31, 32]. Results from these studies have demonstrated the existence of clonal complexes related to host species [32, 33], while no host species correlation was found in isolates from rainbow trout in Swiss farms [34].
MLVA based on VNTRs is another method that has showed promising results as an approach for separation of strains of other fish pathogenic bacteria [35]. In the search for genetic markers that may help to understand the epidemiology of F. psychrophilum, such a MLVA system based on eight VNTRs was established. A total of 53 isolates from different geographical locations, but with the majority obtained in Norway and Chile were used in the analysis. This is the first VNTRs based genotyping system for F. psychrophilum.
The present study shows the same separation between F. psychrophilum isolates from Norway and Chile, as that documented in the previous MLST work [36]. The VNTR method used confirmed also the high diversity of the Chilean isolates shown by MLST, and other work by Avendaño et al. [37] including a large number of isolates. In contrast, the Norwegian strains had low variability, which was also observed in another MLST study based on a large collection of isolates from Nordic countries [28]. The larger variation among F. psychrophilum isolates from Chile compared to the more limited variation seen among Norwegian isolates suggest that the salmon industry in the two countries are exposed to different reservoirs of the bacterium. A major difference between the two countries is that Norway has natural populations of Atlantic salmon and trout, while no salmonids are naturally occurring in Chile, but have been imported from several different countries and continents over several years [38]. The larger variation observed in Chile could be due to transgenerational transmission of F. psychrophilum from the different countries producing embryos from salmonids that are exported to Chile. The Chilean production of salmon eggs, during the expansion of the aquaculture industry, was not enough to meet the requirements of the industry, and a massive import of salmon embryos from North America and Europe continued for many years until the ISA outbreaks in 2007 [38]. This international commerce of embryos has resulted in the import of fish pathogens, such as ISA virus, IPN virus and piscine reovirus, PRV, to Chile [39–41]. Avendaño et al. [37] using MLST on 91 F. psychrophilum Chilean isolates suggested also that this bacterium was introduced from Europe and North America to Chile.
Studies of other bacterial pathogens using both MLST and VNTR typing systems, have been giving contradictory results. In a study on Chlamydia felis including isolates from different geographical origins, the discriminatory power of the MLVA scheme was superior to MLST [42]. However, a study of the human pathogen Vibrio vulnificus, gave the same separation for both MLST and VNTRs typing systems [43] which shows that the results obtained for F. psychrophilum are not unique. There are few reports using VNTRs to explain the geographical distribution of bacterial animal pathogens [44] and even less such studies on fish. The MLVA typing system that was applied to discriminate isolates of the Atlantic cod pathogen Francisella noatunensis subsp. noatunensis, gave a much better resolution compared to a study of housekeeping genes [26, 45]. A later study arrived at the same conclusion when using MLVA compared to PFGE method on a larger data set including 91 isolates of the same bacterium [46].
In the present study the resolution given by the VNTRs could not be used to discriminate among the strains from different fish hosts species with the exception of a cluster of F. psychrophilum collected from Atlantic salmon (clade D, VS4 and VS8). This cluster is formed by several farmed and wild strains from the North to the South of Norway and the pattern could be a result of movements of infected Atlantic salmon embryos and smolts during industrial production. Traditionally all brood fish production sites and the major production of smolts are located in western Norway, which mean that large quantities of salmon embryos and smolts are moved north. Hence, to a certain degree, the same transmission mechanisms as the long distance transmission from the northern to the southern hemisphere could explain the pattern of transmission in Norway. The extensive transmission of F. psychrophilum in connection with farming of salmonids could have blurred a previous existence of host specific strains of this bacterium, ie. a high infection pressure in farming areas could possibly explain presence of Atlantic salmon specific strains in rainbow trout and vice versa. However, the existence of a clonal complex connected to rainbow trout has been suggested to exist in Norway [28]. The existence of host specific strains of other fish bacteria, as Yersinia ruckeri, has also been shown using MLST [47], while no host specificity were observed in a study of 41 isolates from Renibacterium salmoninarum from Atlantic salmon and rainbow trout [27]. Future studies, including a larger number of F. psychrophilum isolates analysed by VNTRs may, however, give a better understanding of the genetic relationships between fish hosts and F. psychrophilum.
There are few reports in the literature concerning the stability of VNTR in bacteria subcultures at different temperatures. A report on F. noatunensis ssp. noatunensis, showed stability on the VNTRs selected loci over 3 years [46], although a previous work on the same pathogen reports instability in one VNTR after passages at different temperatures [26]. Another study of VNTR from C. felis showed high stability after 10 passages [42]. In the present study no variation was observed in a F. psychrophilum isolate after 12 passages at 4 and 15 °C, suggesting that the selected VNTRs could be considered as stable genetic markers.
The present study was not able to separate between isolates with respect to tissue tropism that could have given indications of differences in virulence. Hence, experimental challenge studies, using isolates with different VNTR profiles or MLST, have to be performed with respect to future knowledge of virulence factors and development of efficient vaccines.
Conclusions
The present study tried to clarify aspects of transmission of F. psychrophilum in Chile and Norway using VNTRs as genetic markers, ie. a MLVA system. The method gave, to a large extent, the same resolution as that found in a previous MLST study Apablaza et al. [36], with a high diversity among Chilean isolates and less variation among the Norwegian isolates. The method was not able to separate isolates with respect to host species except for one cluster of distinct isolates from wild Atlantic salmon in Norway. It is not clear if this is due to limited resolution of the method or a result of high infection pressure between infected populations of Atlantic salmon and rainbow trout in Chile and Europe. The potential of the VNTR analysis as a method for separation of isolates with respect to virulence have to be tested in challenge experiments using isolates with different profiles. In addition, future studied should include more isolates of F. psychrophilum for a more detailed study of the resolution power of the two methods, MLST and VNTR analysis.
Methods
Isolates
Collection of F. psychrophilum
The isolates included in this study were obtained from different fish species, countries, and tissues mainly in the period from 2006 to 2012 (Table 1). A table with the information of the majority of isolates is presented in Apablaza et al. [36] and a complete overview of all the isolates is presented in Additional file 3. Fish referred to as trout (Salmo trutta) in this study include brown trout from rivers and sea trout.
This study was performed in strict accordance with the recommendations of the Norwegian Animal Welfare Act (01.01.2010) and the work followed the regulations set by The Norwegian Food Safety Authority. The bacterial samples were obtained from the Atlantic salmon after they had been anaesthetized by a blow to the head and killed by instantly decapitation. This procedure complies with Norwegian fish welfare regulations.
Isolation of F. psychrophilum
The F. psychrophilum isolates were cultured from different tissues using TYES-A added glucose (FLPA) [48]. The Chilean, the Danish and the Scottish isolates were transported in agar tubes to our laboratory. The Norwegian samples were cultured directly from fish tissues at the laboratory of the Fish Diseases Research Group of the University of Bergen. The procedures for culturing and identification of the isolates were explained in a previous work [36]. The agar plates were incubated at 15 °C for 48–72 h up to three weeks. After identification of the bacteria the isolates were preserved by adding glycerol 28 % and stored at −80 °C or in liquid nitrogen. DNA was extracted using a DNeasy kit (Qiagen, Hilden, Dusseldorf, Germany) as described by in the manufacturer’s instructions, and stored at -20 °C. To identify F. psychrophilum a set of specific primers for 16S rRNA gene were used [49]. PCR amplification, visualization, cleaning of PCR products, and sequencing were as described by Apablaza et al. 2013 [36].
The complete genome of F. psychrophilum JIP02/86 strain, obtained from GenBank (Accession number AM398681) was analysed for the presence of potential tandem repeat regions by the software Tandem Repeat Finder (Benson G, 1999). Loci with tandem repeats consisting of four to ten nucleotides were selected for the analysis. Specific oligonucleotide primers for 12 potential variable number tandem repeats (VNTR) loci were designed using the Vector NTI Suite 9.0 program package (InforMax Inc). The oligonucleotide primers had Tm ranging from 52 to 59° (Table 2).
The amplification of VNTRs was performed as described in Brevik et al. [26]. The PCR products were visualized using gel electrophoresis with 1 % agarose. In cases where the initial PCR did not amplify a target sequence, a second or third PCR was run. This included using previous PCR product as template in an identical PCR run (repeated PCR), or using a gradient of different Tm on extracted DNA to allow more specific binding of the primers. Additional pairs of flanking primers were designed for VNTR7-F/R and VNTR 13-F/R, since not all the isolates were amplified using the initial primers (Table 2). The cleaning and sequencing of PCR products were also performed as described in Brevik et al. [26]. Initially the 12 set of primers that identified potentially suitable VNTRs were tested using DNA extracted from 10 isolates from Norway and Chile and the type strain of F. psychrophilum, NCIMB 1947. Among these isolates eight VNTRs showed variation, while no variation was found in the latter four VNTRs. Bases on these results the remaining 42 isolates were tested for the eight former VNTRs only.
VNTR stability
To test the stability in vitro of all the VNTRs at different temperatures the isolate No12-49-As-Op was tested after 12 passages and at incubation temperatures of 4 °C (No12-49-As-Op/4) and 15 °C (No12-49-As-Op/15).
Phylogenetic analysis
The relationships between the type species and the F. psychrophilum isolates from Norway and Chile, based on the variation in the VNTRs, were analysed using neighbour-joining (NJ) distance method and principal component analysis (PCA).
At each VNTR locus in a single taxon, the VNTR was coded as a discrete character (i.e., 1–9, A-H) based on the specific number of repeats at the region in question. These allele profiles were used to construct a data matrix within the Mesquite System for Phylogenetic Analysis (Maddison, W.P. and D.R Maddison), [26].
Principal component analysis
The principal component analysis (PCA) of VNTRs of all the isolates was performed in Unscrambler 9.8, CAMO, Oslo, Norway.
Abbreviations
- BCWD:
-
Bacterial cold water disease
- DNA:
-
Deoxyribonucleic acid
- FLPA:
-
TYES-A added glucose agar
- MLST:
-
Multi locus of sequence typing
- MLVA:
-
Multiple loci VNTR analysis
- NJ:
-
Neighbour-joining distance method
- PCA:
-
Principal component analysis
- PCR:
-
Polymerase chain reaction
- PFGE:
-
Pulsed-field electrophoresis
- RTFS:
-
Rainbow trout fry syndrome
- VNTR:
-
Variable number of tandem repeats
- VS:
-
VNTR set
References
Gliniewicz K, Plant KP, LaPatra SE, LaFrentz BR, Cain K, Snekvik KR, et al. Comparative proteomic analysis of virulent and rifampicin-attenuated Flavobacterium psychrophilum. J Fish Dis. 2012;35(7):529–39.
Henríquez-Nuñez H, Evrard O, Kronvall G, Avendaño-Herrera R. Antimicrobial susceptibility and plasmid profiles of Flavobacterium psychrophilum strains isolated in Chile. Aquaculture. 2012;354:38–44.
Castillo D, Higuera G, Villa M, Middelboe M, Dalsgaard I, Madsen L, et al. Diversity of Flavobacterium psychrophilum and the potential use of its phages for protection against bacterial cold water disease in salmonids. J Fish Dis. 2012;35(3):193–201.
Boutin S, Bernatchez L, Audet C, Derome N. Antagonistic effect of indigenous skin bacteria of brook charr (Salvelinus fontinalis) against Flavobacterium columnare and F. psychrophilum. Vet Microbiol. 2012;155(2–4):355–61.
Nematollahi A, Decostere A, Pasmans F, Haesebrouck F. Flavobacterium psychrophilum infections in salmonid fish. J Fish Dis. 2003;26(10):563–74.
Barnes ME, Brown ML. A review of Flavobacterium psychrophilum biology, clinical signs, and Bacterial Cold Water Disease prevention and treatment. Open Fish Sc J. 2011;4:40–8.
Long A, Call DR, Cain KD. Investigation of the link between broodstock infection, vertical transmission, and prevalence of Flavobacterium psychrophilum in eggs and progeny of rainbow trout and coho salmon. J Aquatic Animal Health. 2014;26(2):66–77.
Sudheesh PS, LaFrentz BR, Call DR, Siems WF, LaPatra SE, Wiens GD, et al. Identification of potential vaccine target antigens by immunoproteomic analysis of a virulent and a non-virulent strain of the fish pathogen Flavobacterium psychrophilum. Dis Aquatic Org. 2007;74(1):37–47.
Nishiki I, Furukawa M, Matui S, Itami T, Nakai T, Yoshida T. Epidemiological study on Lactococcus garvieae isolates from fish in Japan. Fish Sci. 2011;77(3):367–73.
Suanyuk N, Kong FR, Ko D, Gilbert GL, Supamattaya K. Occurrence of rare genotypes of Streptococcus agalactiae in cultured red tilapia Oreochromis sp. and Nile tilapia O. niloticus in Thailand-relationship to human isolates? Aquaculture. 2008;284(1–4):35–40.
Shukla S, Shukla SK, Sharma R, Kumar A. Identification of Mycobacterium species from apparently healthy freshwater aquarium fish using partial sequencing and PCR- RFLP analysis of heat shock protein (hsp65) gene. J Appl Ichthyol. 2014;30(3):513–20.
Erfanmanesh A, Soltani M, Pirali E, Mohammadian S, Taherimirghaed A. Genetic characterization of Streptococcus iniae in diseased farmed rainbow trout (Onchorhynchus mykiss) in Iran. Sci World J. 2012;6.
Steigen A, Karlsbakk E, Plarre H, Watanabe K, Overgard AC, Brevik O, et al. A new intracellular bacterium, Candidatus Similichlamydia labri sp. nov. (Chlamydiaceae) producing epitheliocysts in ballan wrasse, Labrus bergylta (Pisces, Labridae). Arch Microbiol. 2015;197(2):311–8.
Abayneh T, Colquhoun DJ, Sorum H. Multi-locus sequence analysis (MLSA) of Edwardsiella tarda isolates from fish. Vet Microbiol. 2012;158(3–4):367–75.
Zhang XJ, Yang WM, Wu H, Gong XN, Li AH. Multilocus sequence typing revealed a clonal lineage of Aeromonas hydrophila caused motile Aeromonas septicemia outbreaks in pond-cultured cyprinid fish in an epidemic area in central China. Aquaculture. 2014;432:1–6.
Abayneh T, Colquhoun DJ, Austin D, Sørum H. Multilocus variable number tandem repeat analysis of Edwardsiella piscicida isolates pathogenic to fish. J Fish Dis. 2014;37(11):941–8.
Sun G, Chen C, Li J, Gui X, Li X, Chen L, et al. Discriminatory potential of a novel set of variable number of tandem repeats for genotyping Mycobacterium marinum. Vet Microbiol. 2011;152(1–2):200–4.
Torsvik J, Johansson S, Johansen A, Ek J, Minton J, Raeder H, et al. Mutations in the VNTR of the carboxyl-ester lipase gene (CEL) are a rare cause of monogenic diabetes. Hum Genet. 2010;127(1):55–64.
Lindstedt B-A. Genotyping of selected bacterial enteropathogens in Norway. Int J Med Microbiol. 2011;301(8):648–53.
Wang X, Gu W, Cui Z, Luo L, Cheng P, Xiao Y, et al. Multiple-locus variable-number tandem-repeat analysis of pathogenic Yersinia enterocolitica in China. PLoS ONE. 2012;7(5):e37309.
Fujita K, Ito Y, Hirai T, Maekawa K, Imai S, Tatsumi S, et al. Genetic relatedness of Mycobacterium avium-intracellulare complex isolates from patients with pulmonary MAC disease and their residential soils. Clin Microbiol Infection. 2013;19(6):537–41.
Hannou N, Llop P, Faure D, López MM, Moumni M. Characterization of Erwinia amylovora strains from Middle Atlas Mountains in Morocco by PCR based on tandem repeat sequences. Eur J Plant Pathol. 2013;136:655–974.
Skaala Ø, Høyheim B, Glover K, Dahle G. Microsatellite analysis in domesticated and wild Atlantic salmon (Salmo salar L.): allelic diversity and identification of individuals. Aquaculture. 2004;240(1–4):131–43.
Ferrada S, Canales-Aguirre CB, Galleguillos R, Barrera A, Gallardo JA. Development of microsatellite markers in the copepod, Caligus rogercresseyi Boxshall & Bravo, 2000 (Copepoda, Caligidae). Crustaceana. 2011;84(3):375–81.
Broza YY, Danin-Poleg Y, Lerner L, Valinsky L, Broza M, Kashi Y. Epidemiologic study of Vibrio vulnificus infections by using variable number tandem repeats. Emerg Infect Dis. 2009;15(8):1282–5.
Brevik OJ, Ottem KF, Nylund A. Multiple-locus, variable number of tandem repeat analysis (MLVA) of the fish-pathogen Francisella noatunensis. BMC Vet Res. 2011;7.
Matejusova I, Bain N, Colquhoun DJ, Feil EJ, McCarthy U, McLennan D, et al. Multilocus variable-number tandem-repeat genotyping of Renibacterium salmoninarum, a bacterium causing bacterial kidney disease in salmonid fish. BMC Microbiol. 2013;13.
Nilsen H, Sundell K, Duchaud E, Nicolas P, Dalsgaard I, Madsen L, et al. Multilocus sequence typing identifies epidemic clones of Flavobacterium psychrophilum in Nordic countries. Applied Env Microbiol. 2014;80(9):2728–36.
Fujiwara-Nagata E, Ikeda J, Sugahara K, Eguchi M. A novel genotyping technique for distinguishing between Flavobacterium psychrophilum isolates virulent and avirulent to ayu, Plecoglossus altivelis altivelis (Temminck & Schlegel). J Fish Dis. 2012;35(7):471–80.
Hesami S, Allen KJ, Metcalf D, Ostland VE, MacInnes JI, Lumsden JS. Phenotypic and genotypic analysis of Flavobacterium psychrophilum isolates from Ontario salmonids with bacterial coldwater disease. Can J Microbiol. 2008;54(8):619–29.
Sundell K, Wiklund T. Characteristics of epidemic and sporadic Flavobacterium psychrophilum sequence types. Aquaculture. 2015;441:51–6.
Nicolas P, Mondot S, Achaz G, Bouchenot C, Bernardet JF, Duchaud E. Population structure of the fish-pathogenic bacterium Flavobacterium psychrophilum. Applied Env Microbiol. 2008;74(12):3702–9.
Siekoula-Nguedia C, Blanc G, Duchaud E, Calvez S. Genetic diversity of Flavobacterium psychrophilum isolated from rainbow trout in France: Predominance of a clonal complex. Vet Microbiol. 2012;161(1–2):169–78.
Strepparava N, Nicolas P, Wahli T, Segner H, Petrini O. Molecular epidemiology of Flavobacterium psychrophilum from Swiss fish farms. Dis Aquatic Org. 2013;105(3):203–10.
van Belkum A. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA). FEMS Immunol Med Microbiol. 2007;49(1):22–7.
Apablaza P, Løland AD, Brevik ØJ, Ilardi P, Battaglia J, Nylund A. Genetic variation among Flavobacterium psychrophilum isolates from wild and farmed salmonids in Norway and Chile. J Applied Microbiol. 2013;114(4):934–46.
Avendaño-Herrera R, Houel A, Irgang R, Bernardet JF, Godoy M, Nicolas P, et al. Introduction, expansion and coexistence of epidemic Flavobacterium psychrophilum lineages in Chilean fish farms. Vet Microbiol. 2014;170(3–4):298–306.
Sernapesca: Statitics of ovas importation by origin 2008–2013. Servicio Nacional de Pesca. http://sernapesca.cl 2013. (In spanish)
Kibenge MJT, Iwamoto T, Wang Y, Morton A, Godoy MG, Kibenge FSB. Whole-genome analysis of piscine reovirus (PRV) shows PRV represents a new genus in family Reoviridae and its genome segment S1 sequences group it into two separate sub-genotypes. Virol J. 2013;10.
Vike S, Nylund S, Nylund A. ISA virus in Chile: evidence of vertical transmission. Arch Virol. 2009;154(1):1–8.
Mutoloki S, Evensen O. Sequence similarities of the capsid gene of Chilean and European isolates of infectious pancreatic necrosis virus point towards a common origin. J Gen Virol. 2011;92:1721–6.
Laroucau K, Di Francesco A, Vorimore F, Thierry S, Pingret JL, Bertin C, et al. Multilocus variable-number tandem-repeat analysis scheme for Chlamydia felis genotyping: comparison with multilocus sequence typing. J Clin Microbiol. 2012;50(6):1860–6.
Broza YY, Raz N, Lerner L, Danin-Poleg Y, Kashi Y. Genetic diversity of the human pathogen Vibrio vulnificus: A new phylogroup. Int J Food Microbiol. 2012;153(3):436–43.
Russell CL, Smith EM, Calvo-Bado LA, Green LE, Wellington EMH, Medley GF, et al. Multiple locus VNTR analysis highlights that geographical clustering and distribution of Dichelobacter nodosus, the causal agent of footrot in sheep, correlates with inter-country movements. Infect Gen Evol. 2014;22:273–9.
Ottem KF, Nylund A, Karlsbakk E, Friis-Moller A, Kamaishi T. Elevation of Francisella philomiragia subsp noatunensis Mikalsen et al. (2007) to Francisella noatunensis comb. nov syn. Francisella piscicida Ottem et al. (2008) syn. nov. and characterization of Francisella noatunensis subsp. orientalis subsp nov., two important fish pathogens. J Applied Microbiol. 2009;106(4):1231–43.
Duodu S, Wan XH, Tandstad NM, Larsson P, Myrtennas K, Sjodin A, et al. An improved multiple-locus variable-number of tandem repeat analysis (MLVA) for the fish pathogen Francisella noatunensis using capillary electrophoresis. BMC Vet Res. 2013;9.
Bastardo A, Ravelo C, Romalde JL. Multilocus sequence typing reveals high genetic diversity and epidemic population structure for the fish pathogen Yersinia ruckeri. Env Microbiol. 2012;14(8):1888–97.
Cepeda C, Garcia-Marquez S, Santos Y. Improved growth of Flavobacterium psychrophilum using a new culture medium. Aquaculture. 2004;238(1–4):75–82.
Giovannoni SJ. The polymerase chain reduction. In: Stakkebrandt E, Goodfellow M, editors. Modern Microbial Methods: Sequencing and Hybridization Techniques in Bacterial Systematics. Wiley: Chichester; 1991. p. 177––203.
Acknowledgements
To Cermaq Chile, represented by Julio Mendoza for the support of this work. The PhD position connected to this work was financed by the University of Bergen and Nofima, Bergen. Also thanks to T. Harry Birkbeck from University of Glasgow, United Kingdom who kindly provided the two isolates from Scotland (Sc10-47-As-K and Sc-48-Rt-Sp) included in the study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PA was the main contributor for writing the manuscript and main responsible to realize the study, ØJB identified the VNTR loci, contribute to the experimental design and writing. SM was responsible to perform the PCA and writing. SV, PI, and JB contributed with the writing of the manuscript. ID contributed to the critical review of the document and AN coordinated the project, contributed to different parts of project design and reviewed all drafts. All authors are approved the final document.
Additional files
Additional file 1:
Accesion numbers of all the VNTR sequences included in the study.
Additional file 2:
Allelic profile of the 53 F. psychrophilum isolates included in this study.
Additional file 3:
Overview of the isolates of F. psychrophilum included in the study.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Apablaza, P., Brevik, Ø.J., Mjøs, S. et al. Variable Number of Tandem Repeats (VNTR) analysis of Flavobacterium psychrophilum from salmonids in Chile and Norway. BMC Vet Res 11, 150 (2015). https://doi.org/10.1186/s12917-015-0469-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-015-0469-7