Abstract
Background
We performed phenome-wide Mendelian randomization analysis (MR-PheWAS), two-sample MR analysis, and systemic review to comprehensively explore the health effects of milk consumption in the European population.
Methods
Rs4988235 located upstream of the LCT gene was used as the instrumental variable for milk consumption. MR-PheWAS analysis was conducted to map the association of genetically predicted milk consumption with 1081 phenotypes in the UK Biobank study (n=339,197). The associations identified in MR-PheWAS were examined by two-sample MR analysis using data from the FinnGen study (n=260,405) and international consortia. A systematic review of MR studies on milk consumption was further performed.
Results
PheWAS and two-sample MR analyses found robust evidence in support of inverse associations of genetically predicted milk consumption with risk of cataract (odds ratio (OR) per 50 g/day increase in milk consumption, 0.89, 95% confidence interval (CI), 0.84–0.94; p=3.81×10−5), hypercholesterolemia (OR, 0.91, 95% CI 0.86–0.96; p=2.97×10−4), and anal and rectal polyps (OR, 0.85, 95% CI, 0.77–0.94; p=0.001). An inverse association for type 2 diabetes risk (OR, 0.92, 95% CI, 0.86–0.97; p=0.003) was observed in MR analysis based on genetic data with body mass index adjustment but not in the corresponding data without body mass index adjustment. The systematic review additionally found evidence that genetically predicted milk consumption was inversely associated with asthma, hay fever, multiple sclerosis, colorectal cancer, and Alzheimer’s disease, and positively associated with Parkinson’s disease, renal cell carcinoma, metabolic syndrome, overweight, and obesity.
Conclusions
This study suggests several health effects of milk consumption in the European population.
Similar content being viewed by others
Background
As major components of traditional Western diets, milk products that contain many essential nutrients may a play role in human health [1]. A substantial number of studies have examined the association between milk consumption and a wide range of health outcomes, like diabetes [2], cardiovascular disease [3,4,5], and certain cancers [6,7,8], albeit with inconclusive findings. The US dietary guideline recommends at least three servings (237 ml) per day of milk or equivalent portions of cheese, yogurt, or other dairy products for adults and children aged ≥9 years [9]. This standard is substantially higher than the current mean milk consumption (around 1.6 serving/day) among American adults [1] and populations in other parts of the world [10]. However, whether health benefits can be observed for such increased levels of milk consumption to justify the current dietary intake recommendation remains uncertain according to a recent review that scarcely found any well-established associations between milk consumption and disease risk [1]. In addition, most evidence is based on observational studies that are prone to be influenced by methodological limitations, particularly confounding and reverse causality.
Milk consumption is substantially influenced by the lactase gene (LCT), which encodes the enzyme lactase that is essential for lactose digestion. A genetic variant located upstream of the LCT gene was associated with lactase persistence and higher milk consumption in individuals of European descent [11, 12]. Mendelian randomization (MR) is an epidemiological approach that can strengthen causal inference by using one or more genetic variants as instrumental variable for an exposure [13]. Previous MR studies found possible associations of genetically predicted milk consumption with obesity and metabolic syndrome [14,15,16,17,18,19], Alzheimer’s disease [20], multiple sclerosis [20], colorectal [21] and kidney [22] cancers, hay fever [23], and asthma [23]. However, not all potential milk-intake–related outcomes, such as cataract [24], have been examined by MR studies, and further studies are required to replicate and strengthen these findings. Phenome-wide association study (PheWAS) is featured as a hypothesis-free design with the integrality of well-defined and widely adopted phenome framework to generate associations for further examination. To comprehensively explore the health effects of milk consumption, we conducted this study to examine the associations of milk consumption with a wide range of diseases by conducting a PheWAS analysis in the UK Biobank study as well as a two-sample MR analysis where the LCT gene variant was used as proxy for milk consumption. Of note, the UK Biobank study is based on a generally healthy young population which may not be perfectly suitable to study certain outcomes with a low prevalence. Thus, we further conducted a systematic review of published MR studies on milk consumption to complement the findings.
Methods
Study design
Figure 1 shows the study design of the present investigation. We first conducted a MR-phenome-wide association study (MR-PheWAS) to explore the health effects of milk consumption in the UK Biobank study. We then conducted a two-sample MR analysis with data from the FinnGen study and international consortia to replicate the observed associations in the MR-PheWAS. Finally, we conducted a systematic review of MR studies on milk consumption to comprehensively synthesize the evidence to validate any possible health effects.
Genetic instrument selection
A single nucleotide polymorphism (SNP, rs4988235) located upstream of the LCT gene was used as the instrumental variable in the MR-PheWAS and two-sample MR analyses. This SNP showed a strong association with milk consumption in the European populations [11]. One additional milk consumption increasing allele of rs4988235 was associated with an increase of 17.1 (95% confidence interval [CI] 10.6–23.6) g/day milk consumption in a sub-cohort of the European Prospective Investigation into Cancer and Nutrition-InterAct study including 12,722 participants [11]. Summary-level statistics on the rs4988235-milk association (i.e., beta and corresponding standard error coefficients) from the above study were used and rescaled to 50 g/day increase in the current MR analysis. The rs4988235-milk association was also verified in a cohort study comprising 73,715 Danish individuals where each additional milk consumption increasing allele of rs4988235 was associated with an increase of 0.58 (95% CI 0.49–0.68) glasses/week in milk consumption (p=9×10−36) [12]. In this study, rs4988235 explained around 2% phenotypic variance in milk consumption [12]. Rs4988235 explained around 2.5% variance in milk consumption and had a F-statistic of 1055 in the UK Biobank study. The milk consumption increasing allele of rs4988235 also showed a strong inverse association with lactose intolerance in the R6 FinnGen study data (p=1.54×10−61).
MR-PheWAS in UK Biobank
We performed the PheWAS analysis using data on germline genotype and health outcomes from the UK Biobank study including 339,197 unrelated White individuals aged between 40 and 69 years in 2006–2010. The detailed quality control procedures of genotype data and the selection of white population are described in Additional file 1: Supplementary Methods and Fig. S1. Briefly, samples that were identified as a sex mismatch, outliers with high heterozygosity or with high missing rate, putative sex chromosome aneuploidy, individuals with excess relatives, or non-White ancestry were all excluded from the analysis. The largest possible subset of individuals without relatedness was identified using an algorithm implemented in the R package “i-graph (v1.0.1)” developed by Bycroft and colleagues [25]. The PheCODE schema [26] was used to define phenotypes based on an integrative application of 10,750 unique ICD (International Classification of Diseases)-10 codes and 3113 ICD-9 codes with diagnostic information from national medical records (e.g., inpatient hospital episode records, cancer registry, and death registry, Additional file 1: Table S1). Detailed information on genotyping and quality control in UK Biobank has been described in our previous studies [25, 27]. The UK Biobank received ethical permits from the North West Multi-centre Research Ethics Committee, the National Information Governance Board for Health and Social Care in England and Wales, and the Community Health Index Advisory Group in Scotland. All participants provided written informed consent.
Two-sample MR
Two-sample MR analyses were conducted based on data from the R6 FinnGen study including up to 260,405 individuals [28] and international consortia [29,30,31]. The FinnGen study is a growing project combining data on germline genotypes and a wide range of health outcomes from Finnish biobanks and health registries. Detailed information on FinnGen and used international consortia is shown in Additional file 1: Table S2.
Systematic review of MR studies on milk consumption
We further performed a systematic review of MR studies on milk consumption in the PubMed database to complement the associations obtained from the MR-PheWAS and two-sample MR analyses. We searched articles up to 3 March 2022 using the following search strategy: “Mendelian Randomization Analysis” [Mesh] OR mendelian[tiab] AND “milk” [Mesh] OR milk[tiab] (Additional file 1: Table S3). We extracted data on the first author, year of publication, used genetic instrument(s), outcome(s) studied, numbers of cases and controls, and the association estimates in the main statistical analysis. The literature search, review process, and data extraction were done in parallel by two authors (S.Y and Y.L.).
Statistical analysis
In PheWAS, we calculated a weighted genetic score by adding up the number of milk consumption increasing alleles for rs4988235 weighted by its effect size on milk intake. We confined the analysis to outcomes with at least 200 cases [32]. The associations of genetically predicted milk consumption with phenotypes were estimated using logistic regression models adjusted for age, sex, body mass index (BMI), assessment center, and the first ten genetic principal components. We also conducted a sensitivity analysis without adjustment for BMI as well as additional stratification analyses of participants based on non-overweight (BMI <25 kg/m2) and overweight (BMI ≥25 kg/m2) status. The false discovery rate correction with the method by Benjamini-Hochberg was used to account for multiple comparisons in MR-PheWAS analysis [33].
In a two-sample MR analysis, the Wald ratio method was used to estimate the causal association (i.e., the beta coefficient for the effect of the SNP on the outcome divided by the beta coefficient for the effect of the SNP on milk consumption) [34]. The standard error of the ratio estimate is estimated using the delta method [35]. The odds ratio (OR) and corresponding CI were scaled to genetically predicted 50 g/day increase in milk consumption in MR-PheWAS and two-sample MR analyses. The association with a p<0.05 was deemed significant in the two-sample MR analysis. All tests were two-sided and conducted using a R package by Carroll et al [36], and MendelianRandomziation package [37] in R Software 4.0.2.
Results
MR-PheWAS analysis
MR-PheWAS analysis was based on 182,072 females and 157,125 males in the UK biobank. The characteristics of participants are shown in Additional file 1: Table S4. Using the PheCODE schema, we defined 1853 distinct phenotypes. After the removal of outcomes with less than 200 cases, 1081 phenotypes classified into 18 disease categories were included in the analysis (Additional file 1: Table S5). A total of 70 phenotypes were associated with genetically predicted milk consumption at the nominal significance level (p<0.05) (Additional file 1: Table S6). After accounting for multiple testing, genetically predicted higher milk consumption was associated with decreased risk of 8 outcomes, including cataract, type 2 diabetes, diabetes mellitus, disorders of lipoid metabolism hypercholesterolemia, hyperlipidemia, macular degeneration (senile) of retina, and anal and rectal polyp (Table 1 and Fig. 2). The associations were stable in the sensitivity analysis without adjustment for BMI (Table 1). The associations were overall consistent in the analysis by overweight status albeit nonsignificant in the non-overweight population with small numbers of cases (Additional file 1: Table S7).
Results of the phenome-wide MR association analysis on genetically proxied milk consumption for clinical outcomes in the UK Biobank. The Y-axis corresponds to the logarithms of the p values derived from the phenome-wide MR association analyses. The red lines correspond to the statistical significance level (false discovery rate <0.05). Associations surviving the significance criteria are labeled by name
Two-sample MR analysis
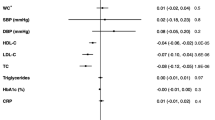
The associations for cataract, lipid metabolism, and anal and rectal polyp were observed in two-sample MR analysis (Table 2). Per 50 g/day increase in genetically predicted milk consumption, the OR was 0.97 (95% CI, 0.95, 0.99; p=0.006) for cataract, 0.91 (95% CI, 0.83, 0.98; p=0.015) for benign neoplasm of transverse colon, and 0.97 (95% CI, 0.94, 1.00; p=0.043) for benign neoplasm of colon. For the same increase in genetically predicted milk consumption, the changes of different lipid biomarkers were −0.015 (95% CI, −0.019, −0.012; p=1.97×10−19) for high-density lipoprotein cholesterol, −0.018 (95% CI, −0.022, −0.015; p=2.96×10−27) for low-density lipoprotein cholesterol, and −0.020 (95% CI, −0.023, −0.017; p=8.29×10−34) for triglycerides. Genetically predicted milk consumption was inversely associated with risk of type 2 diabetes in the analysis based on genome-wide association study data for type 2 diabetes with BMI adjustment (OR, 0.97, 95% CI, 0.96, 0.99; p=0.003), but not in the corresponding analysis based on data without BMI adjustment (Table 2).
Systematic review of MR studies on milk consumption
A total of 80 studies were obtained from the search in the PubMed database and 18 relevant studies were included in the review. Information on 18 studies is present in Additional file 1: Table S8. For disease outcomes, genetically predicted higher milk consumption was associated with a lower risk of asthma, hay fever, multiple sclerosis, colorectal cancer, and Alzheimer’s disease, and a higher risk of Parkinson’s disease, renal cell carcinoma, metabolic syndrome, overweight and obesity (Fig. 3). For biomarkers, genetically predicted higher milk consumption was associated with lower levels high- and low-density lipoprotein cholesterol and higher levels of fasting insulin (Table 3).
Discussion
In this study, MR-PheWAS and two-sample MR analyses found robust evidence in support of inverse associations of genetically predicted milk consumption with risk of cataract, hyperlipidemia, and anal and rectal polyps. An inverse association between genetically predicted milk intake and type 2 diabetes risk was observed in MR analysis based on data with BMI adjustment, but not in the analysis without BMI adjustment. Systematic review of MR studies revealed additional inverse associations of genetically predicted milk consumption with asthma, hay fever, multiple sclerosis, colorectal cancer, Alzheimer’s disease, and blood lipid levels, but positive associations for Parkinson’s disease, renal cell carcinoma, metabolic syndrome, overweight and obesity, and levels of BMI and fasting insulin.
A novel finding of our study is the observed inverse association between milk consumption and risk of cataract. This association has been studied in only a few previous studies with conflicting findings [24]. Current daily milk intake was associated with a reduced risk of cataract extraction in a cross-sectional analysis including 5930 individuals from the US National Health and Nutrition Examination Survey [24]. However, the inverse association with incident cataract was not clearly observed in a cohort analysis of 5860 subjects from the PREvención con DIeta MEDiterránea Study [38]. Notably, the participants in this study were in high cardiovascular risk (e.g., with a high prevalence of diabetes, hypercholesterolemia, and hypertension) [38], which is different from the UK Biobank individuals with a generally good health status. Our recent MR study found causal associations of several cardiovascular risk factors with an increased risk of cataract [39]. In the PREvención con DIeta MEDiterránea Study, participants with at least one cardiovascular risk factor were at a high risk of cataract. Thus, the moderate protective effect of milk consumption on cataract might not be observed in this population.
Previous MR study reported a significant association between milk consumption and colorectal cancer risk [21]; however, this association did not pass multiple corrections in our PheWAS analysis (P=0.039). Instead, we found a significant association between milk consumption and the risk of anal and rectal polyps. Given that colorectal polyp is an important risk factor for colorectal cancer, the observed MR association between milk consumption and anal and rectal polyp partly supported the inverse association between milk consumption and colorectal cancer as reported by previous MR studies [6]. In a meta-analysis of 15 cohort studies with 11,733 incident colorectal cancer patients, higher consumptions of total dairy products and total milk were associated with a lower risk of colorectal cancer [6]. In a recent diet-wide association study for risk of colorectal cancer, one standard deviation increment increase in milk consumption was associated with 5% lower risk of incident colorectal cancer in 396,792 adults from the European Prospective Investigation into Cancer and Nutrition (EPIC) study [40]. In addition, by comparing results for different sites of colorectal polyp, our findings imply that high milk intake may exert more protective effects on transverse colon and possibly on sigmoid colon compared to other sites.
Evidence on the association between milk consumption and risk of type 2 diabetes is inconsistent between observational and MR studies. A review based on 12 meta-analyses found that most studies supported an inverse association between the consumption of total milk, in particular low-fat milk, and risk of incident type 2 diabetes [2]. Nevertheless, this inverse association was not observed in MR studies [11, 16, 18]. The MR analysis using rs4988235 as genetic instrument for milk consumption found no association between milk intake and diabetes risk (OR, 0.99: 95% CI, 0.93, 1.05) in 9686 diabetes cases and 12,134 controls from the EPIC study [11]. Our study found an association between milk consumption and risk of type 2 diabetes in data with BMI adjustment, but not in the analysis without adjustment for BMI. Considering BMI appears to be a collider factor as well as a mediator in the association between milk and type 2 diabetes, the adjustment for BMI was likely to bias the association. Thus, our findings along with previous MR studies [11, 16, 18], do not support a causal association between milk consumption and type 2 diabetes risk.
Several other health outcomes have been linked to milk consumption in previous MR studies. There were inverse associations for asthma [23], hay fever [23], multiple sclerosis [20], and Alzheimer’s disease [20], and positive associations for Parkinson’s disease [20], renal cell carcinoma [22], metabolic syndrome [14], and overweight and obesity [16]. However, these outcomes were not captured by our PheWAS analysis since our study might be lack of power to detect such associations, like for multiple sclerosis, Alzheimer’s disease, and Parkinson’s disease with a low prevalence or smaller number of cases in the UK biobank participants. Future studies with larger sample sizes and independent study populations are required to replicate and validate these reported MR findings. For biomarkers, genetically predicted higher milk intake was associated with lower levels of high- and low-density lipoprotein cholesterol [18] and higher levels of BMI [15, 17,18,19] and fasting insulin [18].
Several mechanisms may explain above identified associations. We observed that milk consumption favored the blood lipid profile, which can partly explain the inverse associations between milk consumption and outcomes with a high level of lipids as a risk factor, such as cataract [41]. In addition, calcium, riboflavin, vitamin D, phosphorous are rich in milk and these nutrients exert various health effects [42, 43]. A clinic-based study found that a high intake of riboflavin was associated with the decreased risk of several cataract-related endpoints [44]. Milk consumption has been associated with profiles of gut microbiome, which may mediate the associations of milk consumption and identified health issues. A genome-wide association study found that lactase gene locus was associated with Bifidobacterium abundance [45] and the colonization of Bifidobacterium animalis markedly reduces the polyp burden and possibly the risk of colorectal cancer [46]. Another study implied that Bifidobacterium possibly mediated the association between increased milk consumption and lower levels of triglycerides [47].
There are several strengths of this study, including a valid genetic instrument for milk consumption [11], a wide range of phenotypes studied, an independent replication analysis using data from other sources, and a comprehensive collection of findings from a systematic review. Several limitations need consideration when interpreting our results. Since the used genetic instrument is strongly associated with milk intake but not with intake of other dairy products, this study could only examine the health effects of total milk intake but not the effects of fermented milk products, such as cheese and yogurt. In addition, we could not differentiate the associations for intake of skimmed and unskimmed milk in this analysis based on summary level data. Our analysis might be challenged by horizontal pleiotropy since the genetic variant used as the proxy for milk intake might be associated with intake of other foods [11]. However, this horizontal pleiotropy should be minimal given the modest associations between rs4988235 and intake of a few other foods [11]. Possible nonlinear associations of milk intake with health outcomes could not be explored due to insufficient statistical power with only one genetic instrument explaining a modest proportion (about 2%) of the variance in milk consumption. We might have overlooked certain weak associations in MR-PheWAS analysis due to an inadequate power caused by a small phenotypic variance explained by the milk intake-associated SNP and a small number of cases in the UK Biobank. Even though an MR-PheWAS analysis based on a larger sample of a meta-analysis of UK Biobank and FinnGen may increase power, this approach cannot be conducted due to a lack of individual-level data in FinnGen. In addition, we could not compare the associations in different units of milk consumption even though it did not hinder the causal inference in these associations.
Conclusions
In summary, this study revealed several health effects of milk consumption in the European population with evidence from a comprehensive investigation based on different study designs. These findings suggest that promoting milk intake may act as a dietary strategy for certain diseases’ prevention, such as for cataract and hyperlipidemia. Future clinical trials are needed to verify our results.
Availability of data and materials
Data from UK Biobank can be obtained via application (https://www.ukbiobank.ac.uk/). The UK Biobank is an open-access resource and bona fide researchers can apply to use the UK Biobank dataset by registering and applying at http://ukbiobank.ac.uk/register-apply/. This research was conducted using the UK Biobank study under Application Number 66354. Data used in two-sample MR analysis and review of MR studies can be obtained by a reasonable request to the corresponding author. Codes for MR-PheWAS can be obtained in https://github.com/xueli157/xueli157/blob/main/PheWAS/PheWAS%20Function_R%20script.txt
Abbreviations
- BMI:
-
Body mass index
- CI:
-
Confidence interval
- ICD:
-
International Classification of Diseases
- MR:
-
Mendelian randomization
- MR-PheWAS:
-
Phenome-wide Mendelian randomization analysis
- OR:
-
Odds ratio
- SNP:
-
Single nucleotide polymorphism
References
Willett WC, Ludwig DS. Milk and Health. N Engl J Med. 2020;382(7):644–54.
Alvarez-Bueno C, Cavero-Redondo I, Martinez-Vizcaino V, Sotos-Prieto M, Ruiz JR, Gil A. Effects of Milk and Dairy Product Consumption on Type 2 Diabetes: Overview of Systematic Reviews and Meta-Analyses. Adv Nutr. 2019;10(suppl_2):S154–s163.
Fontecha J, Calvo MV, Juarez M, Gil A, Martínez-Vizcaino V. Milk and Dairy Product Consumption and Cardiovascular Diseases: An Overview of Systematic Reviews and Meta-Analyses. Adv Nutr. 2019;10(suppl_2):S164–s189.
Chen Z, Ahmed M, Ha V, Jefferson K, Malik V, Ribeiro PAB, et al. Dairy Product Consumption and Cardiovascular Health: a Systematic Review and Meta-Analysis of Prospective Cohort Studies. Adv Nutr. 2021.
Larsson SC, Crippa A, Orsini N, Wolk A, Michaëlsson K. Milk Consumption and Mortality from All Causes, Cardiovascular Disease, and Cancer: A Systematic Review and Meta-Analysis. Nutrients. 2015;7(9):7749–63.
Barrubés L, Babio N, Becerra-Tomás N, Rosique-Esteban N, Salas-Salvadó J. Association Between Dairy Product Consumption and Colorectal Cancer Risk in Adults: A Systematic Review and Meta-Analysis of Epidemiologic Studies. Adv Nutr. 2019;10(suppl_2):S190–s211.
Bermejo LM, López-Plaza B, Santurino C, Cavero-Redondo I, Gómez-Candela C. Milk and Dairy Product Consumption and Bladder Cancer Risk: A Systematic Review and Meta-Analysis of Observational Studies. Adv Nutr. 2019;10(suppl_2):S224–s238.
López-Plaza B, Bermejo LM, Santurino C, Cavero-Redondo I, Álvarez-Bueno C, Gómez-Candela C. Milk and Dairy Product Consumption and Prostate Cancer Risk and Mortality: An Overview of Systematic Reviews and Meta-analyses. Adv Nutr. 2019;10(suppl_2):S212–s223.
McGuire S. Scientific Report of the 2015 Dietary Guidelines Advisory Committee. Washington, DC: US Departments of Agriculture and Health and Human Services, 2015. Adv Nutr. 2016;7(1):202–4.
Gateway to dairy production and products [https://www.fao.org/dairy-production-products/products/en/].
Vissers LET, Sluijs I, van der Schouw YT, Forouhi NG, Imamura F, Burgess S, et al. Dairy Product Intake and Risk of Type 2 Diabetes in EPIC-InterAct: A Mendelian Randomization Study. Diabetes Care. 2019;42(4):568–75.
Bergholdt HKM, Larsen MK, Varbo A, Nordestgaard BG, Ellervik C. Lactase persistence, milk intake, hip fracture and bone mineral density: a study of 97 811 Danish individuals and a meta-analysis. J Intern Med. 2018;284(3):254–69.
Burgess S, Thompson SG. Mendelian Randomization: Methods for Using Genetic Variants in Causal Estimation. London: Chapman and Hall/CRC; 2015.
Almon R, Alvarez-Leon EE, Engfeldt P, Serra-Majem L, Magnuson A, Nilsson TK. Associations between lactase persistence and the metabolic syndrome in a cross-sectional study in the Canary Islands. Eur J Nutr. 2010;49(3):141–6.
Almon R, Álvarez-León EE, Serra-Majem L. Association of the European lactase persistence variant (LCT-13910 C>T polymorphism) with obesity in the Canary Islands. PLoS One. 2012;7(8):e43978.
Bergholdt HK, Nordestgaard BG, Ellervik C. Milk intake is not associated with low risk of diabetes or overweight-obesity: a Mendelian randomization study in 97,811 Danish individuals. Am J Clin Nutr. 2015;102(2):487–96.
Hartwig FP, Horta BL, Smith GD, de Mola CL, Victora CG. Association of lactase persistence genotype with milk consumption, obesity and blood pressure: a Mendelian randomization study in the 1982 Pelotas (Brazil) Birth Cohort, with a systematic review and meta-analysis. Int J Epidemiol. 2016;45(5):1573–87.
Yang Q, Lin SL, Au Yeung SL, Kwok MK, Xu L, Leung GM, et al. Genetically predicted milk consumption and bone health, ischemic heart disease and type 2 diabetes: a Mendelian randomization study. Eur J Clin Nutr. 2017;71(8):1008–12.
Vimaleswaran KS, Zhou A, Cavadino A, Hyppönen E. Evidence for a causal association between milk intake and cardiometabolic disease outcomes using a two-sample Mendelian Randomization analysis in up to 1,904,220 individuals. Int J Obes. 2021;45(8):1751–62.
Zhang Z, Wang M, Yuan S, Larsson SC, Liu X. Genetically Predicted Milk Intake and Risk of Neurodegenerative Diseases. Nutrients. 2021;13(8).
Larsson SC, Mason AM, Kar S, Vithayathil M, Carter P, Baron JA, et al. Genetically proxied milk consumption and risk of colorectal, bladder, breast, and prostate cancer: a two-sample Mendelian randomization study. BMC Med. 2020;18(1):370.
Timpson NJ, Brennan P, Gaborieau V, Moore L, Zaridze D, Matveev V, et al. Can lactase persistence genotype be used to reassess the relationship between renal cell carcinoma and milk drinking? Potentials and problems in the application of Mendelian randomization. Cancer Epidemiol Biomark Prev. 2010;19(5):1341–8.
Skaaby T, Kilpeläinen TO, Mahendran Y, Huang LO, Sallis H, Thuesen BH, et al. Association of milk intake with hay fever, asthma, and lung function: a Mendelian randomization analysis. Eur J Epidemiol. 2022.
Mustafa OM, Daoud YJ. Is Dietary Milk Intake Associated with Cataract Extraction History in Older Adults? An Analysis from the US Population. J Ophthalmol. 2020;2020:2562875.
Yuan S, Wang L, Sun J, Yu L, Zhou X, Yang J, et al. Genetically predicted sex hormone levels and health outcomes: phenome-wide Mendelian randomization investigation. Int J Epidemiol. 2022.
Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–10.
Li X, Meng X, He Y, Spiliopoulou A, Timofeeva M, Wei WQ, et al. Genetically determined serum urate levels and cardiovascular and other diseases in UK Biobank cohort: A phenome-wide mendelian randomization study. PLoS Med. 2019;16(10):e1002937.
Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner K, et al. FinnGen: Unique genetic insights from combining isolated population and national health register data. medRxiv. 2022.
Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505–13.
Chiou J, Geusz RJ, Okino ML, Han JY, Miller M, Melton R, et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature. 2021;594(7863):398–402.
Graham SE, Clarke SL, Wu KH, Kanoni S, Zajac GJM, Ramdas S, et al. The power of genetic diversity in genome-wide association studies of lipids. Nature. 2021;600(7890):675–9.
Verma A, Bradford Y, Dudek S, Lucas AM, Verma SS, Pendergrass SA, et al. A simulation study investigating power estimates in phenome-wide association studies. BMC Bioinformatics. 2018;19(1):120.
Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Methodol. 1995;57(1):289–300.
Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26(5):2333–55.
Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–65.
Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30(16):2375–6.
Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734–9.
Camacho-Barcia L, Bulló M, García-Gavilán JF, Martínez-González MA, Corella D, Estruch R, et al. Dairy products intake and the risk of incident cataracts surgery in an elderly Mediterranean population: results from the PREDIMED study. Eur J Nutr. 2019;58(2):619–27.
Yuan S, Wolk A, Larsson SC. Metabolic and lifestyle factors in relation to senile cataract: a Mendelian randomization study. Sci Rep. 2022;12(1):409.
Papadimitriou N, Bouras E, van den Brandt PA, Muller DC, Papadopoulou A, Heath AK, et al. A prospective diet-wide association study for risk of colorectal cancer in EPIC. Clin Gastroenterol Hepatol. 2022;20(4):864–873.e13.
Hiller R, Sperduto RD, Reed GF, D'Agostino RB, Wilson PW. Serum lipids and age-related lens opacities: a longitudinal investigation: the Framingham Studies. Ophthalmology. 2003;110(3):578–83.
Yuan S, Yu L, Gou W, Wang L, Sun J, Li D, et al. Health effects of high serum calcium levels: Updated phenome-wide Mendelian randomisation investigation and review of Mendelian randomisation studies. EBioMedicine. 2022;76:103865.
Thakur K, Tomar SK, Singh AK, Mandal S, Arora S. Riboflavin and health: A review of recent human research. Crit Rev Food Sci Nutr. 2017;57(17):3650–60.
Glaser TS, Doss LE, Shih G, Nigam D, Sperduto RD, Ferris FL 3rd, et al. The Association of Dietary Lutein plus Zeaxanthin and B Vitamins with Cataracts in the Age-Related Eye Disease Study: AREDS Report No. 37. Ophthalmology. 2015;122(7):1471–9.
Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2021;53(2):156–65.
Liao W, Khan I, Huang G, Chen S, Liu L, Leong WK, et al. Bifidobacterium animalis: the missing link for the cancer-preventive effect of Gynostemma pentaphyllum. Gut Microbes. 2021;13(1):1847629.
Shuai M, Zuo LS, Miao Z, Gou W, Xu F, Jiang Z, et al. Multi-omics analyses reveal relationships among dairy consumption, gut microbiota and cardiometabolic health. EBioMedicine. 2021;66:103284.
Acknowledgements
We want to acknowledge the participants and investigators of the FinnGen study.
Funding
Open access funding provided by Uppsala University. This study was supported by research grants from the Swedish Heart Lung Foundation (Hjärt-Lungfonden, 20210351), the Swedish Research Council (Vetenskapsrådet, 2019-00977), the Swedish Research Council for Health, Working Life and Welfare (Forte; 2018-00123), and the Swedish Cancer Society (Cancerfonden). XL is supported by the Natural Science Fund for Distinguished Young Scholars of Zhejiang Province (LR22H260001) and the National Nature Science Foundation of China (82204019).
Author information
Authors and Affiliations
Contributions
S.Y., X.L., L.Q.Q., and S.C.L. have made substantial contributions to the conception and design of the work. S.Y. and X.L have made substantial contributions to the data acquisition and analysis. S.Y. wrote the first draft of the manuscript. S.Y., J.S., Y.L., F.X., D.L., F.J., Z.W., X.L., L.Q.Q., and S.C.L. have made substantial contributions to the interpretation of data and critical revisions of the manuscript for important intellectual content. S.Y., J.S., Y.L., F.X., D.L., F.J., Z.W., X.L., L.Q.Q., and S.C.L. have read and approved the submitted version of the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The UK Biobank received ethical permits from the North West Multi-centre Research Ethics Committee, the National Information Governance Board for Health and Social Care in England and Wales, and the Community Health Index Advisory Group in Scotland (REC reference: 21/NW/0157). The FinnGen study was proved by the Coordinating Ethics Committee of the Helsinki and Uusimaa Hospital District (Nr HUS/990/2017). All participants provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Methods
. Table S1. Mappings of ICD-10 and ICD-9 codes to the phenotypes identified by MR-PheWAS at the nominal significance level (p<0.05). Table S2. Information on the FinnGen study and international consortia. Table S3. Search strategy in the PubMed database. TableS4. Characteristics of participants in the UK Biobank (N=339,197). Table S5. Outcomes included in the analyses and outcomes excluded due to power (N<200 cases). Table S6. Phenotypes associated with genetically predicted milk consumption in MR-PheWAS at the nominal significance level (p<0.05) among unrelated white British sample (N=339,197). Table S7. Phenotypes associated with genetically proxied milk consumption by overweight status in MR-PheWAS analysis in the UK Biobank. Table S8. Information on included studies in review. Figure S1. Flow diagram of quality control procedures and the selection of target population.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yuan, S., Sun, J., Lu, Y. et al. Health effects of milk consumption: phenome-wide Mendelian randomization study. BMC Med 20, 455 (2022). https://doi.org/10.1186/s12916-022-02658-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12916-022-02658-w