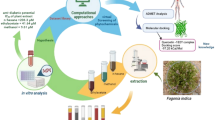
Abstract
Background
The dried bark of Ailanthus altissima (Mill.) Swingle is widely used in traditional Chinese medicine for the treatment of ulcerative colitis. The objective of this study was to explore the therapeutic basis of the dried bark of Ailanthus altissima (Mill.) Swingle for the treatment of ulcerative colitis based on Virtual Screening–Molecular Docking–Activity Evaluation technology.
Methods
By searching the Traditional Chinese Medicine Systems Pharmacology TCMSP Database and Analysis Platform, 89 compounds were obtained from the chemical components of the dried bark of Ailanthus altissima (Mill.) Swingle. Then, after preliminarily screening the compounds based on Lipinski’s rule of five and other relevant conditions, the AutoDock Vina molecular docking software was used to evaluate the affinity of the compounds to ulcerative colitis-related target proteins and their binding modes through use of the scoring function to identify the best candidate compounds. Further verification of the compound’s properties was achieved through in vitro experiments.
Results
Twenty-two compounds obtained from the secondary screening were molecularly docked with ulcerative colitis-related target proteins (IL-1R, TLR, EGFR, TGFR, and Wnt) using AutoDock Vina. The free energies of the highest scoring compounds binding to the active cavity of human IL-1R, TLR, EGFR, TGFR, and Wnt proteins were − 8.7, − 8.0, − 9.2, − 7.7, and − 8.5 kcal/mol, respectively. The potential compounds, dehydrocrebanine, ailanthone, and kaempferol, were obtained through scoring function and docking mode analysis. Furthermore, the potential compound ailanthone (1, 3, and 10 µM) was found to have no significant effect on cell proliferation, though at 10 µM it reduced the level of pro-inflammatory factors caused by lipopolysaccharide.
Conclusion
Among the active components of the dried bark of Ailanthus altissima (Mill.) Swingle, ailanthone plays a major role in its anti-inflammatory properties. The present study shows that ailanthone has advantages in cell proliferation and in inhibiting of inflammation, but further animal research is needed to confirm its pharmaceutical potential.
Similar content being viewed by others
Background
Ulcerative colitis (UC), a nonspecific inflammatory bowel disease with an unknown etiology [1], often occurs in the colon and rectum and is diagnosed by colonoscopy [2, 3]. Patients with recurrent and prolonged UC are prone to cancer [4, 5], and UC has been classified as a refractory disease by the World Health Organization (WHO) [6]. Western medicine does not effectively control UC flares, and cannot prevent its recurrence [7]. In contrast to the long treatment cycle [8], adverse drug effects [9], high treatment costs [10, 11], and other disadvantages of Western medicine, traditional Chinese Medicine (TCM) has significant effects in the clinical treatment of UC [12].
The bark of Ailanthus altissima (Mill.) Swingle (BAA), which is also known as chun root bark, citrus white bark, ailanthus bark, and kucha bark, is commonly used in TCM. It has a bitter, astringent, and cold taste [13]. BAA can clear heat, dry dampness, astringe the intestines, and stop bleeding, and it has been used to treat chronic dysentery, enteritis, diarrhea, gastric and duodenal ulcers, blood in the stool, nocturnal emission, leucorrhea, and other diseases [14, 15]. Modern medical research has found that BAA and its preparations have a good curative effect on UC [13, 16,17,18]. A 100%-pure decoction of BAA has been shown to have inhibitory effects on Shigella flexneri, Shigella sohnii, Bacillus typhi, and Escherichia coli in vitro, and Staphylococcus aureus is moderately sensitive to it [19, 20]. BAA extract can also reduce the activity of nitric oxide (NO) free radicals and nitric oxide synthase (NOS) [18, 21], which are inflammatory factors associated with UC [22].
Molecular docking technology docks known small-molecule active substances with related target proteins, and can elucidate the mechanism of action between active TCM ingredients and targets at the molecular level [23, 24]. At present, molecular docking technology has played an important role in elucidating the mechanism of action between active TCM ingredients and related target proteins in many diseases, such as cardiovascular disease [25] and cancer [26]. Research on the treatment of UC is in the initial stage, and the pharmacodynamic substances that are effective against UC have not yet been fully investigated [27]. The molecular docking virtual screening technology used to screen the pharmacodynamic substances (active component groups) of BAA that are effective in the treatment of UC is relatively simple and can obtain accurate results. Various signaling pathways that are closely related to UC, including TLR/IL-1R, EGFR, TGFR and Wnt/β-catenin. Research has found that the TLR/IL-1R signaling pathways play a crucial role in host defense and inflammation [28], the EGFR and Wnt signaling pathways are closely related to the repair of colonic mucosal injury [29, 30], TGFR can also significantly promote the apoptosis of T cells [31], thereby exacerbating the development of UC [32]. Therefore, TLR, IL-1R, EGFR, TGFR, and Wnt can serve as docking proteins.
The aim of this study was to identify all of the known chemical components of BAA using the Traditional Chinese Medicine Systems Pharmacology (TCMSP) Database and Analysis Platform, combined with the five principles of drug-like molecules (Lipinski rule) and standard conditions, such as oral bioavailability and drug similarity, to preliminarily screen for compounds that could be developed into therapeutics. We set human interleukin-1 receptor (IL-1R), toll-like receptor (TLR), epidermal growth factor receptor (EGFR), transforming growth factor receptor (TGFR), and Wnt proteins as the targets of UC, and AutoDock Vina molecular docking software was used to sort the bioactive compounds of BAA by scoring function. Additionally, the primary-screening compound group and key targets of disease pathogenesis were also explored. The affinity and binding mode of the proteins were then used to screen for active ingredients with potential therapeutic effectiveness against UC. In addition, potential compounds were further tested in vitro.
Materials and methods
Construction of ligand library (compound)
The TCMSP Database and Analysis Platform were used to establish a preliminary-screening compound group for TCM (https://old.tcmsp-e.com/tcmsp.php) [33]. The small-molecule data were downloaded in mol2 format from the TCMSP database according to the small molecule MOLID number. The small molecules were then imported into ChemBio3D Ultra 14.0 (Cambridgesoft, Cambridge, MA, USA) for energy minimization. The minimum RMS gradient was set at 0.001, and the small molecule was saved in mol2 format. The optimized small molecule was imported into AutoDockTools (Scripps Research Institute, La Jolla, CA, USA) for hydrogenation, and the charge was calculated and assigned, in addition to the rotatable bond being set. The file was saved in “pdbqt” format.
Preparation of receptors (proteins)
UC-related target protein data came from Protein Data Bank (PDB, http://www.rcsb.org/) [34]. The most relevant UC target proteins, including human IL-1R, TLR, EGFR, TGFR, and Wnt, were set as the receptor proteins. High resolution was preferred, and the original ligand had high structural similarity to the active ingredient to be docked. Pymol (DeLano Scientific LLC, Palo Alto, CA, USA) was used to remove protein crystal water and original ligands. The protein structure was imported into AutoDockTools (Scripps Research Institute, La Jolla, CA, USA) for hydrogenation, and the charge was calculated and assigned. The type of atom was specified, and the file was saved in “pdbqt” format.
Docking settings
AutoDock Vina [35] (Scripps Research Institute, La Jolla, CA, USA) was used for docking, and the EGFR [36](PDB ID: 7LGS) target-related parameters were set as follows: center_x = 29.720,center_y = − 19.100, center_z = 5.833, and size of the grid box = 60 × 60 × 60 (the spacing between each grid point was 0.375Å).The rest of the parameters were the default settings. IL-1R (PDB ID: 5R8J) target-related parameters were set as follows: center_x = 38.545, center_y = 13.206, center_z = 68.747, and size of the grid box = 114 × 94 × 110 (the spacing between each grid point was 0.375Å); the rest of the parameters were the default settings. TLR1 (PDB ID: 7NT7) target-related parameters were set as follows: center_x = 38.062, center_y = 15.372, center_z = − 6.727, and size of the grid box was set to 84 × 106 × 120 (the spacing between each grid point was 0.375 Å).The rest of the parameters were the default settings. The Wnt-8 (PDB ID: 4FOA) target-related parameters were set as follows: center_x = − 62.066, center_y = − 9.0, center_z = 9.934, and size of the grid box = 126 × 126 × 126 (the spacing between each grid point was 0.375Å); the rest of the parameters were the default settings. TGFR (PDB ID: 5Q1L) target-related parameters were set as follows: center_x = 4.108, center_y = 9.252, center_z = 7.215, and size of the grid box was set to 60 × 60 × 60 (the spacing between each grid point was 0.375Å).The rest of the parameters were the default settings.
Molecular docking between chemical components and target proteins
The compounds with the highest docking score for each protein were selected to visualize the docking results using Discovery Studio (Accelrys Software Inc., San Diego, USA).
CCK-8 cytotoxicity assay
Human normal colonic epithelial NCM460 cells and mouse RAW264.7 macrophages were provided by the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). We took the cells in the logarithmic growth phase (NCM460; RAW264.7), counted them, pipetted 200 µL of cell suspension into a 96-well plate, and set the cell density to 5 × 103 cells per well. We divided the samples into blank and control groups, which were both placed in an incubator for routine culture for 24 h. We accurately weighed the test drug ailanthone, used DMSO to prepare a mother solution with a concentration of 200 µM, and then sequentially diluted it with DMSO to concentrations of 1, 3, 10, 30, and 50 µM for administration. The next day, in both the blank group and the control group, the medium was replaced with a fresh culture medium. Specifically, in the administration group, a culture medium containing drugs (1, 3, 10, 30, and 50 µM) was used, while we added 1 µL of DMSO to the solvent control group; afterward they were placed in an incubator for routine culture. After 24 h, 10 µL of CCK-8 solution was added to each well, and the cells were placed in an incubator for further incubation for 2.5 h. We measured the absorbance value A of each well at a wavelength of 450 nm with an enzyme-linked immunosorbent assay, calculated the average value of the absorbance of each group, and calculated the inhibition rate according to the following formula: Inhibition rate% = 100% × [(A control group − A administration group)/(A control group − A blank group)]. The IC50 values of drugs on cells (NCM460; RAW264.7) were calculated with GraphPad Prism software.
Determination of anti-inflammatory activity
According to the CCK-8 cytotoxicity test, the concentration of the drug whose toxicity to Raw264.7 cells was less than the IC50 value was selected for anti-inflammatory activity detection, and drug concentrations of 1, 3, and 10 µM were selected. Raw264.7 cells in the logarithmic growth phase were taken and trypsinized; the cells were collected and centrifuged at 1000 rpm/min; the supernatant was discarded, and the cells were resuspended in a fresh culture medium. We counted the cells, adjusted the cell density, pipetted 100 µL of the cell suspension into a 24-well plate, and then added 700 µL of the culture medium to achieve a cell density of 1 × 105 cells per well. We set the blank group and the control group and placed them in an incubator for routine culture for 24 h. The test drug ailanthone was precisely weighed, and the mother solution was prepared with DMSO to achieve a concentration of 200 µM, which was sequentially diluted with DMSO to concentrations of 1, 3, and 10 µM for administration. The DMSO content of the administered cells was less than or equal to 1%. The cells were first incubated with drugs for 2 h, then exposed to LPS with a final concentration of 1 µg/mL, and finally co-cultured for 24 h. After 24 h, the cell supernatant of each experimental group was collected, and the absorbance value was detected by a microplate reader in accordance with the operation instructions of the ELISA kit. The contents of tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), and interleukin-1β (IL-1β) in the supernatant were determined.
Statistical analysis
The statistical software used was GraphPad Prism 8 (La Jolla, CA, USA), and P < 0.05 indicated a statistically significant difference.
Results
Preliminary screening of compounds in BAA
Compounds were searched with BAA as the keyword, and 89 small-molecule chemical components were retrieved from the TCMSP database. Eighty-nine compound libraries were initially constructed. The five principles of drug-like and other related screening conditions were used for secondary screening: (1) molecular weight (MW) less than 500; (2) lipid–water partition coefficient (ALogP) less than 5; (3) number of hydrogen-bond donors (Hdon) less than 5; (4) number of hydrogen-bond receptors (Hacc) less than 10; (5) oral bioavailability (OB) greater than 50; and (6) drug similarity (DL) greater than 0.18. The secondary screening resulted in 22 compounds. As shown in Table 1, these 22 compounds were suggested to have good drug-like properties, and were saved in mol2 format for future use.
Scoring function ranking of secondary-screening chemical components and target proteins
The 22 compounds obtained from the secondary screening were molecularly docked with human IL-1R, TLR, EGFR, TGFR, and Wnt proteins using AutoDock Vina software. The binding conformation was scored using a scoring function based on shape matching and energy matching. The docking results of the compounds and receptors were sorted in descending order of the total score, and the results are shown in Table 2. The free energies of binding to the active cavities of human IL-1R, TLR, EGFR, TGFR, and Wnt protein of the highest scoring compound obtained by AutoDock Vina software were − 8.7, − 8.0, − 9.2, − 7.7, and − 8.5 kcal/mol, respectively. A larger absolute value of the binding constant indicates that lower free energy is required for the compound to bind. It is generally believed that when the absolute value is greater than 7, the combination of the compound and the protein is more likely.
Binding pattern of the highest scoring compound and target protein
Figure 1 shows that the compounds bind closely to the surface of human IL-1R, TLR, EGFR, TGFR, and Wnt proteins, and there is a good spatial match between them. All of the compounds bind to the active space of the corresponding protein.
Binding pattern of the highest scoring compounds to human EGFR, IL-1R, TGFR, TLR, and Wnt8 proteins. The colored curly structure represents the amino acid spatial sequence of the target protein, and the yellow structure in the protein cavity represents small molecules. The free energies of binding to the active cavities of human EGFR, IL-1R, TGFR, TLR1 and Wnt proteins of the highest scoring compounds dehydrocrebanine (006315), ailanthone (006284), kaempferol (000422), ailanthone (006284), and ailanthone (006284) are − 8.7, − 8.0, − 9.2, − 7.7, and − 8.5 kcal/mol, respectively
Microscopic binding of the highest scoring compound to amino acid residues near the active site of the target protein
Figure 2 shows that dehydrocrebanine has a noncovalent bond with the residues between the amino acid motifs of threonine (THR) 857 near the active site of human EGFR protein, and ailanthone interacts with the ASP active site of human IL-1R protein. Partic acid (86 amino acid residues) has hydrogen-bonding interactions. Kaempferol forms a cation–π interaction with lysine (LYS) 232 at the active site of human TGFR protein, and ailanthone forms a π–alkyl interaction with tryptophan (TYR) 691 near the active site of TLR. Ailanthone forms a π–alkyl interaction with the LYS 210 active site of human Wnt protein, forming hydrogen-bond interactions.
Microscopic binding of the highest scoring compound to amino acid residues near the active site of the target protein. Green circles and lines represent conventional hydrogen bonds; light blue circles and lines represent carbon hydrocarbon bonds; orange circles and lines represent π–cation interactions; pink circles and lines represent alkyls; and light pink circles and lines represent π–alkyl interactions. The letter and the number under the amino acid represent the abbreviated name and the primary structure of theamino acid sequence
Inhibitory effect of ailanthone on cell proliferatio
Ailanthone was found to be able to combine with L-1R, TLR1, and Wnt at the same time through virtual screening of front molecule docking. It is worth noting that the docking scores were high. However, the computer results could not replace the experiment. The cytotoxicity of ailanthone was tested on cells by CCK-8 experiment (NCM460; RAW264.7). The results showed that cell proliferation was not significantly affected at the concentrations of 1, 3, and 10 µmol of ailanthone (Fig. 3). This safe dose can be used to observe the anti-inflammatory activity of ailanthone.
Ailanthone can inhibit the activity of TNF-α, IL-6, and IL-1β
The activity of TNF-α, IL-6, and IL-1β is closely related to the development of UC. The concentrations of 1, 3, and 10 µM of ailanthone were used to observe the effect on the activity of TNF-α, IL-6, and IL-1β in RAW264.7 cells (Fig. 4). The levels of these pro-inflammatory cytokines decreased after administration of ailanthone (10 µM).
Discussion
In recent years, there have been many studies on the treatment of UC with TCM [37]. Many components in TCM, such as Patrinia scabiosaefolia Fisch [38], Lindera aggregata (Sims) Kosterm [39], Glycyrrhizae [40], Angelica sinensis [41], and BAA [13], have been reported to have therapeutic effects. Particularly significant among these components are flavonoids, glycosides, phenols, polysaccharides, and alkaloids. BAA, a common herbal medicine used in the TCM treatment of UC, has attracted increasing attention. Modern medical research has shown that BAA and its preparations have a good curative effect on UC [13]. At present, research on the treatment of UC is in the initial stage, and its pharmacodynamic substances have not been fully investigated [42]. The molecular docking virtual screening technology used to screen the pharmacodynamic substances (active component groups) of BAA in the treatment of UC is relatively simple and can obtain more accurate results.
Molecular docking technology docks known small-molecule active substances with related target proteins, which can clarify the mechanism of action between active TCM ingredients and targets at the molecular level [43, 44]. At present, molecular docking technology plays an important role in elucidating the mechanism of action between active ingredients of TCM [45, 46] and related target proteins in many diseases, such as cardiovascular disease [25] and cancer [26]. Therefore, we conducted preliminary screening with reference to the five principles of drug-like standards and other screening conditions of “drug similarity” and obtained 22 compounds. The 22 compounds obtained from the secondary screening were molecularly docked with human IL-1R, TLR, EGFR, TGFR, and Wnt proteins using AutoDock Vina software, and the binding conformation was scored using a scoring function. The free energies of the highest scoring compounds binding to the active cavity of human IL-1R, TLR, EGFR, TGFR, and Wnt proteins were − 8.7, − 8.0, − 9.2, − 7.7, and − 8.5 kcal/mol, respectively. It is generally believed that when the absolute value is greater than 7 kcal/mol, the possibility of the combination of the compound and the protein is high [47]. The larger the absolute value of the binding constant, the lower the free energy required for the compound to bind [48]. The compound with the highest function score can be selected in order to construct a compound–protein binding diagram, which can visually convey the binding mode of the active compound with human IL-1R, TLR, EGFR, TGFR, and Wnt proteins and their interaction with the surrounding amino acid residues. It is worth noting that ailanthone was found to be able to combine with L-1R, TLR1, and Wnt proteins at the same time through virtual screening of front molecule docking, and the binding score was very high. This compound was further tested in vitro. Ailanthone had no significant inhibitory effect on cell proliferation within the concentration range of 1–10 µM, and a 10-µM concentration significantly inhibited the pro-inflammatory response induced by LPSa.
Conclusions
Briefly, in this study, the effective substances of BAA in treating UC were studied based on Virtual Screening–Molecular Docking–Activity Evaluation technology. We found that ailanthone had good activity in vitro. Although further animal experiments are needed for verification, ailanthone has a high possibility of becoming an effective drug in the future.
Availability of data and materials
The data used to support the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- BAA:
-
Bark of Ailanthus altissima (Mill.) swingle
- DMSO:
-
Dimethyl sulfoxide
- EGFR:
-
Epidermal growth factor receptor
- IL-1β:
-
Interleukin-1β
- IL-1R:
-
Interleukin-1 receptor
- IL-6:
-
Interleukin-6
- IC50 :
-
Half maximal inhibitory concentration
- LPS:
-
Lipopolysaccharide
- NO:
-
Nitric oxide
- NOS:
-
Nitric oxide synthase
- PDB:
-
Protein data bank
- TCM:
-
Traditional Chinese medicine
- TCMSP:
-
Traditional Chinese Medicine Systems Pharmacology
- TGFR:
-
Transforming growth factor receptor
- TLR:
-
Toll-like receptor
- TNF-α:
-
Tumor necrosis factor-α
- UC:
-
Ulcerative colitis
References
Kikut J, Konecka N, Ziętek M, Kulpa D, Szczuko M. Diet supporting therapy for inflammatory bowel diseases. Eur J Nutr. 2021;60(5):2275–91.
Huang YF, Zhou JT, Qu C, Dou YX, Huang QH, Lin ZX, Xian YF, Xie JH, Xie YL, Lai XP, et al. Anti-inflammatory effects of Brucea javanica oil emulsion by suppressing NF-κB activation on dextran sulfate sodium-induced ulcerative colitis in mice. J Ethnopharmacol. 2017;198:389–98.
Głąbska D, Guzek D, Zakrzewska P, Włodarek D, Lech G. Lycopene, lutein and zeaxanthin may reduce faecal blood, mucus and pus but not abdominal pain in individuals with ulcerative colitis. Nutrients. 2016;8(10):613.
Olen O, Erichsen R, Sachs MC, Pedersen L, Halfvarson J, Askling J, Ekbom A, Sorensen HT, Ludvigsson JF. Colorectal cancer in ulcerative colitis: a scandinavian population-based cohort study. Lancet. 2020;395(10218):123–31.
Li W, Zhao T, Wu D, Li J, Wang M, Sun Y, Hou S. Colorectal Cancer in Ulcerative Colitis: mechanisms, Surveillance and Chemoprevention. Curr Oncol. 2022;29(9):6091–114.
Liu Y, Li BG, Su YH, Zhao RX, Song P, Li H, Cui XH, Gao HM, Zhai RX, Fu XJ, et al. Potential activity of traditional chinese medicine against ulcerative colitis: a review. J Ethnopharmacol. 2022;289:115084.
Li MY, Luo HJ, Wu X, Liu YH, Gan YX, Xu N, Zhang YM, Zhang SH, Zhou CL, Su ZR, et al. Anti-inflammatory Effects of Huangqin Decoction on Dextran Sulfate Sodium-Induced Ulcerative Colitis in mice through regulation of the gut microbiota and suppression of the Ras-PI3K-Akt-HIF-1alpha and NF-kappaB pathways. Front Pharmacol. 2019;10:1552.
Yashiro M. Ulcerative colitis-associated colorectal cancer. World J Gastroenterol. 2014;20(44):16389–97.
Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel J-F. Ulcerative colitis. Lancet. 2017;389(10080):1756–70.
Null KD, Xu YH, Pasquale MK, Su CY, Marren A, Harnett J, Mardekian J, Manuchehri A, Healey P. Ulcerative colitis treatment patterns and cost of care. Value in Health. 2017;20(6):752–61.
Alulis S, Vadstrup K, Olsen J, Jørgensen TR, Qvist N, Munkholm P, Borsi A. The cost burden of Crohn’s disease and ulcerative colitis depending on biologic treatment status - a danish register-based study. BMC Health Serv Res. 2021;21(1):836.
Chen MJ, Ding YX, Tong ZQ. Efficacy and safety of sophora flavescens (Kushen) based traditional chinese medicine in the treatment of ulcerative colitis: clinical evidence and potential mechanisms. Front Pharmacol. 2020;11:603476.
Li X, Li Y, Ma SB, Zhao QQ, Wu JS, Duan LR, Xie YH, Wang SW. Traditional uses, phytochemistry, and pharmacology of Ailanthus altissima (Mill.) Swingle bark: a comprehensive review. J Ethnopharmacol. 2021;275:114121.
Wang RX, Xu Q, Liu L, Liang XJ, Cheng LY, Zhang ML, Shi QW. Antitumour activity of 2-dihydroailanthone from the bark of Ailanthus altissima against U251. Pharm Biol. 2016;54(9):1641–8.
Tan QW, Ni JC, Shi JT, Zhu JX, Chen QJ. Two novel quassinoid glycosides with antiviral activity from the samara of Ailanthus altissima. Molecules. 2020;25(23):5679.
Cho SK, Jeong M, Jang DS, Choi JH. Anti-inflammatory Effects of Canthin-6-one alkaloids from Ailanthus altissima. Planta Med. 2018;84(8):527–35.
Kim HM, Lee JS, Sezirahiga J, Kwon J, Jeong M, Lee D, Choi JH, Jang DS. A New Canthinone-Type Alkaloid Isolated from Ailanthus altissima Swingle. Molecules 2016, 21(5).
Kim HM, Kim SJ, Kim HY, Ryu B, Kwak H, Hur J, Choi JH, Jang DS. Constituents of the stem barks of Ailanthus altissima and their potential to inhibit LPS-induced nitric oxide production. Bioorg Med Chem Lett. 2015;25(5):1017–20.
Wang RX, Lu YJ, Li H, Sun LX, Yang N, Zhao MZ, Zhang ML, Shi QW. Antitumor activity of the Ailanthus altissima bark phytochemical ailanthone against breast cancer MCF-7 cells. Oncol Lett. 2018;15(4):6022–8.
Okunade AL, Bikoff RE, Casper SJ, Oksman A, Goldberg DE, Lewis WH. Antiplasmodial activity of extracts and quassinoids isolated from seedlings ofAilanthus altissima (Simaroubaceae). Phytother Res. 2003;17(6):675–7.
Kim SR, Park Y, Li M, Kim YK, Lee S, Son SY, Lee S, Lee JS, Lee CH, Park HH, et al. Anti-inflammatory effect of Ailanthus altissima (Mill.) Swingle leaves in lipopolysaccharide-stimulated astrocytes. J Ethnopharmacol. 2022;286:114258.
Li J, Tian H, Jiang HJ, Han B. Interleukin-17 SNPs and serum levels increase ulcerative colitis risk: a meta-analysis. World J Gastroenterol. 2014;20(42):15899–909.
Karthick V, Ramanathan K. Virtual screening for oseltamivir-resistant a (H5N1) influenza neuraminidase from traditional chinese medicine database: a combined molecular docking with molecular dynamics approach. Springerplus. 2013;2(1):115.
Wang W, Wan MH, Liao DJ, Peng GL, Xu X, Yin WQ, Guo GX, Jiang FN, Zhong WD, He JX. Identification of potent chloride intracellular channel protein 1 inhibitors from traditional chinese medicine through structure-based virtual screening and molecular dynamics analysis. Biomed Res Int. 2017;2017:4751780.
Maryam A, Khalid RR, Siddiqi AR, Ece A. E-pharmacophore based virtual screening for identification of dual specific PDE5A and PDE3A inhibitors as potential leads against cardiovascular diseases. J Biomol Struct Dynamics. 2021;39(7):2302–17.
Opo FADM, Rahman MM, Ahammad F, Ahmed I, Bhuiyan MA, Asiri AM. Structure based pharmacophore modeling, virtual screening, molecular docking and ADMET approaches for identification of natural anti-cancer agents targeting XIAP protein. Sci Rep. 2021;11(1):4049.
Lv Q, Xing Y, Liu YJ, Chen QZ, Xu JY, Hu LH, Zhang YN. Didymin switches M1-like toward M2-like macrophage to ameliorate ulcerative colitis via fatty acid oxidation. Pharmacol Res. 2021;169:105613.
Wu H, Li XM, Wang JR, Gan WJ, Jiang FQ, Liu Y, Zhang XD, He XS, Zhao YY, Lu XX, et al. NUR77 exerts a protective effect against inflammatory bowel disease by negatively regulating the TRAF6/TLR-IL-1R signalling axis. J Pathol. 2016;238(3):457–69.
Wang Y, Zou J, Jia Y, Zhang X, Wang C, Shi Y, Guo D, Wu Z, Wang F. The mechanism of lavender essential oil in the treatment of Acute Colitis based on “Quantity-Effect” Weight Coefficient Network Pharmacology. Front Pharmacol. 2021;12:644140.
Das S, Feng Q, Balasubramanian I, Lin X, Liu H, Pellon-Cardenas O, Yu S, Zhang X, Liu Y, Wei Z et al. Colonic healing requires Wnt produced by epithelium as well as Tagln + and Acta2 + stromal cells. Development 2022, 149(1).
Hodge SJ, Hodge GL, Reynolds PN, Scicchitano R, Holmes M. Increased production of TGF-beta and apoptosis of T lymphocytes isolated from peripheral blood in COPD. Am J Physiol Lung Cell Mol Physiol. 2003;285(2):L492–499.
Schmitt H, Ulmschneider J, Billmeier U, Vieth M, Scarozza P, Sonnewald S, Reid S, Atreya I, Rath T, Zundler S, et al. The TLR9 agonist Cobitolimod induces IL10-Producing Wound Healing Macrophages and Regulatory T cells in Ulcerative Colitis. J Crohns Colitis. 2020;14(4):508–24.
Tang H, Qin N, Rao C, Zhu J, Wang H, Hu G. Screening of Potential Anti-Thrombotic Ingredients from Salvia miltiorrhiza in Zebrafish and by Molecular Docking. Molecules 2021, 26(22).
Almalki FA, Abdalla AN, Shawky AM, El Hassab MA, Gouda AM. In Silico Approach Using Free Software to Optimize the Antiproliferative Activity and Predict the Potential Mechanism of Action of Pyrrolizine-Based Schiff Bases. Molecules 2021, 26(13).
Zulkipli NN, Zakaria R, Long I, Abdullah SF, Muhammad EF, Wahab HA, Sasongko TH. In Silico Analyses and Cytotoxicity Study of Asiaticoside and Asiatic Acid from Malaysian Plant as Potential mTOR Inhibitors. Molecules 2020, 25(17).
Li X, Huang R, Li M, Zhu Z, Chen Z, Cui L, Luo H, Luo L. Parthenolide inhibits the growth of non-small cell lung cancer by targeting epidermal growth factor receptor. Cancer Cell Int. 2020;20(1):561.
Liu JQ, Liu J, Tong XL, Peng WJ, Wei SS, Sun TL, Wang YK, Zhang BK, Li WQ. Network pharmacology prediction and molecular docking-based strategy to discover the potential pharmacological mechanism of huai hua san against ulcerative colitis. Drug Des Devel Ther. 2021;15:3255–76.
Cho Ej, Shin JS, Noh YS, Cho YW, Hong SJ, Park JH, Lee JY, Lee JY, Lee KT. Anti-inflammatory effects of methanol extract of Patrinia scabiosaefolia in mice with ulcerative colitis. J Ethnopharmacol. 2011;136(3):428–35.
Lai HM, Yang ZB, Lou ZH, Li F, Xie F, Pan W, Xu C, Zhang LL, Zhang S, Zhang LJ, et al. Root extract of Lindera aggregata (Sims) Kosterm. Modulates the Th17/Treg balance to attenuate DSS-induced colitis in mice by IL-6/STAT3 signaling pathway. Front Pharmacol. 2021;12:615506.
Jeon YD, Bang KS, Shin MK, Lee JH, Chang YN, Jin JS. Regulatory effects of glycyrrhizae radix extract on DSS-induced ulcerative colitis. BMC Complement Altern Med. 2016;16(1):459.
Cheng F, Zhang Y, Li Q, Zeng F, Wang KP. Inhibition of dextran sodium sulfate-induced experimental colitis in mice by Angelica Sinensis polysaccharide. J Med Food. 2020;23(6):584–92.
Bartoszek A, Makaro A, Bartoszek A, Kordek R, Fichna J, Salaga M. Walnut Oil Alleviates Intestinal Inflammation and Restores Intestinal Barrier Function in Mice. Nutrients 2020, 12(5).
Fan YH, Liu W, Jin Y, Hou X, Zhang XW, Pan HD, Lu H, Guo XJ. Integrated molecular docking with network pharmacology to reveal the molecular mechanism of simiao powder in the treatment of acute gouty arthritis. Evid Based Complement Alternat Med 2021, 2021:5570968.
Rahman N, Muhammad I, Gul EN, Khan H, Aschner M, Filosa R, Daglia M. Molecular Docking of Isolated Alkaloids for Possible alpha-Glucosidase Inhibition. Biomolecules 2019, 9(10).
Xiang C, Liao Y, Chen Z, Xiao B, Zhao Z, Li A, Xia Y, Wang P, Li H, Xiao T. Network Pharmacology and Molecular Docking to elucidate the potential mechanism of Ligusticum Chuanxiong against Osteoarthritis. Front Pharmacol. 2022;13:854215.
Zhu K, Zhang M, Long J, Zhang S, Luo H. Elucidating the mechanism of action of Salvia miltiorrhiza for the treatment of Acute Pancreatitis based on Network Pharmacology and Molecular Docking Technology. Comput Math Methods Med. 2021;2021:8323661.
Babiaka SB, Simoben CV, Abuga KO, Mbah JA, Karpoormath R, Ongarora D, Mugo H, Monya E, Cho-Ngwa F, Sippl W et al. Alkaloids with Anti-Onchocercal Activity from Voacanga africana Stapf (Apocynaceae): Identification and Molecular Modeling. Molecules 2020, 26(1).
Pan Y, Lu Z, Li C, Qi R, Chang H, Han L, Han W. Molecular Dockings and Molecular Dynamics Simulations reveal the potency of different inhibitors against Xanthine Oxidase. ACS Omega. 2021;6(17):11639–49.
Acknowledgements
The authors would like to thank the staff of the School of Life Sciences, Northwest University, Xi ’an, China.
Funding
This work was supported by the Science and Technology Innovation Project of Shaanxi Province in China (No. 2015SF2-08-01), a grant from the Social Development of Shaanxi Province Key Project (Nos. 2017ZDXM-SF-019 and S2018-ZC-GCZXXY-SF-0005), and the Shaanxi Key Laboratory of Biomedicine (No. 2018SZS41).
Author information
Authors and Affiliations
Contributions
Conceptualization, Shan-bo Ma; Methodology, Lun Liu; Software, Xiang Li; Supervision and Validation, Yan-hua Xie; Writing-original draft, Xiao-peng Shi; Writing-review and editing, Si-wang Wang. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
As this study does not involve animal and humant experiments, the ethical approval and consent to participate are not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ma, Sb., Liu, L., Li, X. et al. Virtual screening–molecular docking–activity evaluation of Ailanthus altissima (Mill.) swingle bark in the treatment of ulcerative colitis. BMC Complement Med Ther 23, 197 (2023). https://doi.org/10.1186/s12906-023-03991-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-023-03991-0