Abstract
Background
Tanshinone I (Tan-I), an ingredient of Salvia miltiorrhiza, displays protective effects in several disease models. We aim to study the effect of Tan-I on renal fibrosis and explore its underlining mechanism.
Methods
Rat renal fibroblasts (NRK-49F) were used as an in vitro model to study the effect of Tan-I. Mouse renal fibrosis model was induced by unilateral ureteral obstruction (UUO) or peritoneally injection of aristolochic acid I (AAI).
Results
We found that Tan-I dose-dependently inhibited the expression of pro-fibrotic markers in rat renal fibroblasts. Masson staining and Western blotting analysis showed that Tan-I treatment attenuated renal fibrosis in UUO or AAI induced fibrotic kidneys. RNA sequencing analysis identified inhibin beta-A (INHBA), a ligand of TGF-β superfamily, as a downstream target of Tan-I in fibrotic kidneys, which were further verified by qPCR. Western blotting analysis showed that INHBA is up-regulated in UUO or AAI induced fibrotic kidneys and Tan-I reduced the expression of INHBA in fibrotic kidneys. Inhibition of INHBA by Tan-I was further confirmed in rat fibroblasts. Moreover, knockdown of INHBA reduced the expression of pro-fibrotic markers and abolished the ani-fibrotic effect of Tan-I in rat renal fibroblasts.
Conclusions
We conclude that Tan-I attenuates fibrosis in fibrotic kidneys through inhibition of INHBA.
Similar content being viewed by others
Background
Chronic kidney disease (CKD) is a common human disease and epidemiological studies show that the prevalence of CKD in adults is as high as more than 10%, resulting in a huge social and economic burden [1]. Renal interstitial fibrosis is the common pathway and main pathological basis for the progression of various CKDs to end-stage renal disease [2]. Pathological characterization of renal interstitial fibrosis is the excessive deposition of extracellular matrix (ECM) in the kidney, which is due to the persistence of pathogenic causes and the imbalance of damage and repair in the inflammatory process leading to secretion of a variety of pro-fibrotic factors such as transforming growth factor beta (TGF-β) [2, 3].
TGF-β is one of the most important factors leading to the development of renal fibrosis, and Smad3 is an important downstream molecule of TGF-β signaling pathway [2, 4, 5]. Inhibition or knockout of Smad3 reduces renal fibrosis in diabetic nephropathy and ureteral obstruction nephropathy [6, 7]. Through activation of Smad pathway, TGF-β promotes the occurrence of epithelial mesenchymal transition (EMT), which is featured by expression of mesenchymal cell markers such as N-cadherin, fibronectin (FN) and α smooth muscle actin (α-SMA) [4]. In fibrotic kidneys, only partial EMT occurs which means renal tubular epithelial cells do not fully transdifferentiate into interstitial fibroblasts, but still integrate into renal tubules. Partial transdifferentiated renal tubular cells produce pro-fibrotic factors and thus promote renal fibrosis progression [4, 8]. Snail is an important transcriptional factor triggering partial EMT and promotes renal fibrosis [8].
Tanshinone I (Tan-I) is a fat-soluble compound extracted from Salvia miltiorrhiza [9]. The effect of Tan-I was extensively studied in the field of cancer biology [10,11,12]. Tan-I inhibits the growth and metastasis of osteosarcoma through inhibition of JAK/STAT3 signaling pathway [12]. Tan-I exerts inhibitory effect on tumor growth through inducing apoptosis and enhancing autophagy of ovarian cancer cells [11]. It has also been reported that Tan-I enhances the radio-sensitivity of lung cancer cells [10]. The protective effect of Tan-I in other diseases has also been shown. Tan-I induces apoptosis of activated hepatic stellate cells suggesting that Tan-I is a potential novel agent for treatment of hepatic fibrosis [13]. It has been shown that Tan-I protects against lung inflammation by activating the Nrf2-dependent antioxidant response [14]. Studies of Tan-I in renal disease is limited. The renal protective effect of Tan-I has been reported in aristolochic acid-induced acute renal injury in rats through induction of hepatic cyp1a [15]. Whether Tan-I is anti-fibrotic in renal disease is yet to be determined.
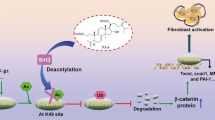
Inhibin beta-A (INHBA), a ligand of TGF-β superfamily, acts as a subunit of inhibin A (activin A) to exert a variety of biological functions and can promote the differentiation of embryonic cells during early embryogenesis [16, 17]. Silencing of INHBA inactivates the TGF-β signaling pathway [18]. INHBA can also promote EMT by activating the TGF-β signaling pathway [19]. The role of INHBA in renal fibrosis is currently unclear.
The aim of current study is to investigate the effect of Tan-I on renal fibrosis and its downstream mechanisms.
Methods
Animal studies
Male C57 mice (C57bl/6j background, SPF grade, 20–25 g) were bought from B&K Universal Group Limited (Shanghai, China) and were housed in the animal facility of Shanghai University of Traditional Chinese Medicine according to local regulations and guidelines. The animal study was performed in accordance with ARRIVE guidelines. Animal experiments were approved by the ethic committee of Shanghai University of Traditional Chinese Medicine (PZSHUTCM18111601).
Mouse were anesthetized with 0.8% sodium pentobarbital (50 mg/kg body weight, i.p.) before UUO operation. UUO model was established by ligation of the left ureter as described previously [20, 21]. Mice were randomly divided into four groups: 1) Sham/DMSO (n = 9), 2) Sham/Tan-I (n = 8), 3) UUO/DMSO (n = 8), and 4) UUO/Tan-I (n = 11) group. Sham or UUO mice were treated with 20 μl DMSO or equivalent volume of Tan-I (50 mg/kg, Topscience, T2907, Shanghai, China) per day by intraperitoneal (i.p.) injection for 14 days starting from day0. Mice were anesthetized with sodium pentobarbital (50 mg/kg body weight, i.p.) and then executed at day14. The dose of Tan-I was taken based on two previous animal studies [15, 22].
To induce aristolochic acid nephropathy (AAN) in mice, aristolochic acid I (AAI, A9451, Sigma; 5 mg/kg body weight) was peritoneally injected two times at day 0 and day7. Mice were randomly divided into three groups: 1) Normal saline (NS)/DMSO (n = 8), 2) AAI/DMSO (n = 8), 3) AAI/Tan-I (n = 8). Mice were treated with 20 μl DMSO or Tan-I daily start from day 14 for two weeks at day 28, and were anesthetized with sodium pentobarbital (50 mg/kg body weight, i.p.) and then executed. Finally, mice were euthanized by cervical dislocation under anesthesia. Kidney tissues were collected.
Cell culture
Normal rat kidney 49F (NRK-49F) cells, a rat kidney interstitial fibroblast cell line, were purchased from national infrastructure of cell line resource, Chinese academy of medical sciences. NRK-49F cells were cultured in DMEM/F12 medium containing 10% fetal bovine serum and 0.5% penicillin and streptomycin in an atmosphere of 5% CO2 and 95% air at 37 °C according to previous studies [20, 21]. To study the anti-fibrotic effect of Tan I, NRK-49F cells were seeded in 6-well plates to 40–50% confluence and were starved with DMEM/F12 medium containing 0.5% fetal bovine serum overnight before the experiment. The next day, cell were refreshed with 0.5%medium and then were exposed to different concentrations of Tan-I (0.5 μM, 5 μM and 50 μM) (Topscience, T2907, Shanghai, China) for 24 h. Protein was extracted from cell lystates for further analysis.
Nonsense control (NC) or rat INHBA siRNA were transfected by Lipofectamine 2000 (11,668–027; Invitrogen) in NRK-49F cells using DMEM/F12 medium containing 10% fetal bovine serum according to the manufacturer’s instruction. Protein was extracted from cell lysates at 24 h after transfection. In another experiment, cells were exposed to vehicle or 50 μM Tan-I for another 24 h. The NC siRNA sequences were as follows: forward, 5′-UUCUCCGAACGUGUCACGUTT-3′; and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′. The rat INHBA siRNA sequences were as follows: forward, 5′-GGGAUGAGGCCGAGGAAAUTT-3′; and reverse, 5′-AUUUCCUCGGCCUCAUCCCTT-3′.
Quantitative real-time PCR and RNA sequence (RNA-seq)
Total RNA was extracted using Trizol (R401-01, Vazyme, Nanjing, China) from kidney samples according to the manufacture’s instruction, which was reverse transcribed to cDNA by Takara PrimeScript RT reagent kit (RR0036A, Kyoto, Japan). The primer sequences for quantitative Polymerase Chain Reaction (qPCR) were listed as follows: mouse INHBA forward, 5'-ATCATCACCTTTGCCGAGTCA-3'; mouse INHBA reverse, 5'-TTCTGCACGCTCCACTACTGA-3'; mouse GAPDH forward, 5'-AGGTCGGTGTGAACGGATTTG-3'; mouse GAPDH reverse, 5'-TGTAGACCATGTAGTTGAGGTCA-3'.
For RNA-seq, kidney samples were prepared from UUO/DMSO and UUO/Tan-I groups with three biological replicates. Total RNA was extracted from the kidney samples using TRIzol® Reagent with removal of genomic DNA by DNase I (TaKara). RNA quality was determined by 2100 Bioanalyser (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). Only qualified RNA samples (OD260/280 = 1.8 ~ 2.2, OD260/230 ≥ 2.0, RIN ≥ 6.5, 28S:18S ≥ 1.0, > 1 μg) were further used for Illumina sequencing. The RNA purification, reverse transcription, library construction, and sequencing were performed at Majorbio Bio-pharm Biotechnology Co., Ltd (Shanghai, China) using Illumina HiSeq X10 (Illumina, San Diego, CA) according to the manufacturer’s instructions. All identified sequences were mapped with Gene Ontology Terms (GO, http://geneontology.org/) to determine the functional and biological properties. To identify DEGs (differential expression genes) between two different samples, the expression level of each transcript was calculated according to the fragments per kilobase of exon per million mapped reads method. Gene abundances was quantified by RSEM (RNA-Seq by Expectation–Maximization). In addition, to identify which DEGs were significantly enriched in metabolic pathways functional-enrichment analyses including Kyoto encyclopedia of genes and genomes (KEGG) were performed at Bonferroni-corrected P value ≤ 0.05 compared with the whole-transcriptome background. The data from this study was deposited in NCBI Sequence Read Archive under accession SRA: SRP347578.
Western blotting analysis
As described previously [20, 21], protein from cells or mouse kidneys was extracted using RIPA lysis buffer (Beyotime Biotech) and measured by the bicinchoninic acid assay before being dissolved in laemmli sample buffer. Samples were subjected to SDS-PAGE gels and were electro-transferred to a PVDF membrane (Merck, Darmstadt, Germany), which was incubated in the blocking buffer (5% nonfat milk, 20 mM Tris HCl, 150 mM NaCl, pH 8.0, 0.1% Tween 20) for 1 h at room temperature. After that membrane was incubated with antibodies for fibronectin (FN; 1:1000, ab23750; Abcam, Cambridge, MA, USA), phosphorylated Smad3(1:1000, ET1609-41, Hangzhou, China, HUABIO, ɑ smooth muscle actin (ɑ-SMA; 1:1000, ET1607-53 Hangzhou, China, HUABIO), SNAIL(A11794, 1:1000, Wuhan, China, Abclonal), inhibin β A(INHBA, 1:1000, ER1911-46, Hangzhou, China, HUABIO; or 1:1000, A5232, Wuhan, China, Abclonal), ɑ-tubulin (1:1000, AF0001; Beyotime Biotech), and GAPDH (1:1000, 60,004–1-lg; Proteintech, Wuhan, Hubei, China) overnight at 4 °C. Binding of the primary antibody was detected by an ECL method (BeyoECL Star, P0018A; Beyotime Biotech) using horseradish peroxidase–conjugated secondary antibodies (goat anti-rabbit IgG, 1:1000, A0208; Beyotime Biotech; or goat anti-mouse IgG, 1:1000, A0216; Beyotime Biotech). The quantification of protein expression was performed by using -Image J (National Institutes of Health). For each figure, all detected proteins including the housekeeper protein were derived from the same membrane.
Masson’s trichrome staining and quantification
Following the protocols described in a previous study [20, 21], mouse kidneys were fixed in 4% paraformaldehyde and further embedded in paraffin. Four-μm thick sections of paraffin-embedded kidney tissue was stained with hematoxylin, and then with ponceau red liquid dye acid complex, which was followed by incubation with phosphomolybdic acid solution. Finally, the tissue was stained with aniline blue liquid and acetic acid. Images were obtained by a microscope (Nikon 80i, Tokyo, Japan). Quantification of collagen positive area was performed by using the ImageJ software. The color threshold (the Hue was set to “125–220”; the saturation was “0–255” and the brightness was “150–225”) was set up to measure the blue positive area of collagen fibers staining. The total area was measured under the threshold mode “0–205”. Ten fields of view at 200 × magnification were captured from each pathological section clockwise, which covered at least 70% of the sample. The blue positive area was divided by the total area for each field, and the average value was calculated for each section.
Statistical analysis
Results were presented as mean ± standard deviation (SD). The data were processed by using IBM SPSS Stastic 22.0 and GraphPad Prism version 8.0 for Windows (GraphPad Software, San Diego, California, United States). The normal distribution of the data was analyzed by Shapiro–Wilk test. When data were normally distributed, they were compared by unpaired student t-test to calculate significance between two groups. Non-normally distributed data were compared using non-parametric tests. A p value of lower than 0.05 was considered statistically significant.
Results
Tan-I inhibits fibrotic changes of rat renal fibroblasts
The anti-fibrotic effect of Tan-I was tested in rat renal fibroblasts (NRK-49F). Treatment with Tan-I for 24 h reduced the phosphorylation of Smad3 in a dose-dependent manner starting from 0.5 μM or 50 μM (Fig. 1). Moreover, the expression of EMT markers such as FN, Snail and α-SMA were reduced by Tan-I in NRK-49F cells (Fig. 1).
Tanshinone I (Tan-I) inhibits fibrotic changes of rat renal fibroblasts. Rat renal fibroblasts (NRK-49F) were starved for 24 h and followed by 24 h treatment with different concentration (0.5 μM, 5 μM, 50 μM) of Tan-I. The expression of fibronectin (FN),α-smooth muscle actin (α-SMA), Snail and phosphorylation of Smad3 (pSmad3) were analyzed by Western blotting and then quantified. Data represents mean ± SD. #p < 0.05 vs DMSO. ##p < 0.01 vs DMSO. N = 6 in each experimental group and one representative result of at least three independent experiments is shown. Full-length blots are presented in Supplementary Fig. 1
Tan-I mitigates renal fibrosis in UUO mice
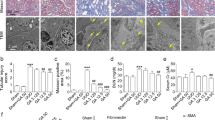
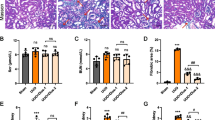
The anti-fibrotic effect of Tan-I was further tested in the mouse unilateral ureteral obstruction (UUO) model, a well-established model of progressive renal fibrosis (Fig. 2A) [23]. Masson trichrome staining revealed massive collagen deposition in UUO kidneys, which was significantly reduced by Tan-I (Fig. 2B). Western blotting further showed a significant up-regulation of FN, Snail, α-SMA and pSmad3 in UUO kidneys as compared with sham kidneys (Fig. 2C). Two weeks treatment with Tan-I significantly reduced the expression of FN, Snail, α-SMA and pSmad3 in UUO kidneys (Fig. 2C).
Tan-I mitigates renal fibrosis in UUO mice. A Study design. Wide type C57 mice received sham or UUO operation and followed by 14 days treatment with Tan-I. Mice were sacrificed at day 14. B Renal fibrosis was assessed by Masson’s trichrome staining and then quantified. Bars = 100 μm. C The expression of FN, α-SMA, Snail and pSmad3 were analyzed by Western blotting and then quantified. N = 8–11 in each experimental group and one representative of at least three independent experiments is shown. Data represents mean ± SD. *p < 0.05 vs Sham/DMSO. **p < 0.01 vs Sham/DMSO. ***p < 0.001 vs Sham/DMSO #p < 0.05 vs UUO/DMSO. ## p < 0.01 vs UUO/DMSO. ### p < 0.001 vs UUO/DMSO. Full-length blots are presented in Supplementary Fig. 2
Tan-I ameliorates renal fibrosis in AAN mice
Mouse aristolochic acid nephropathy (AAN) is a model to study chronic kidney disease (Fig. 3A) [24]. In our study, AAN was induced by two times peritoneally injection of aristolochic acid I (AAI). At 28 days after the first injection of AAI, a strong deposition of collagen was observed in AAN kidneys by Masson staining and two weeks treatment with Tan-I significantly reduced the Masson positive areas in AAN kidneys (Fig. 3B). Similarly, a significant up-regulation of FN, Snail, α-SMA and pSmad3 was observed in AAN kidneys which were reduced by Tan-I treatment (Fig. 3C).
Tan-I ameliorates renal fibrosis in AAN mice. A Study design. Wide type C57 mice received two times peritoneally injection of aristolochic acid I (AAI) to induce aristolochic acid nephropathy (AAN). Mice were treated with Tan-I for two weeks from day14 and were sacrificed at day 28. B Renal fibrosis was assessed by Masson’s trichrome staining and then quantified. Bars = 100 μm. C The expression of FN, α-SMA, Snail1 and pSmad3 were analyzed by Western blotting and then quantified. N = 8 in each experimental group and one representative of at least three independent experiments is shown. Data represents mean ± SD. *p < 0.05 vs normal saline (NS)/DMSO. **p < 0.01 vs NS/DMSO. ***p < 0.001 vs NS/DMSO #p < 0.05 vs AAN/DMSO. ##p < 0.01 vs AAN/DMSO. ###p < 0.001 vs AAN/DMSO. Full-length blots are presented in Supplementary Fig. 3
Tan-I reduces INHBA expression in vitro and in vivo
RNA sequence (RNA-seq) was performed to analyze the differential expressed genes between UUO/DMSO group and UUO/Tan-I group. Differential gene expression analysis revealed that 54 genes were up-regulated and 34 genes were down-regulated in Tan-I treated UUO kidneys (Fig. 4A), among which two genes (INHBA and BMP7) involved in TGF-β signaling pathway were identified by the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Fig. 4B). RNA-seq analysis showed that the fold change of INHBA between UUO/DMSO group and UUO/Tan-I group was 0.297 (P value was 9.87669E-07). The fold change of BMP7 between UUO/DMSO group and UUO/Tan-I group was 2.455 (P value was 1.12E-04). However, we found no significant difference of BMP7 expression between UUO/DMSO and UUO/Tan-I groups by qPCR (data not shown). Down-regulation of INHBA gene in Tan-I treated UUO kidneys was confirmed by qPCR (Fig. 4C). Western blotting analysis showed that Tan-I dose-dependently the expression of INHBA in NRK-49F cells starting from 0.5 μM to 50 μM (Fig. 4D). INHBA was up-regulated in UUO kidneys as compared to sham kidneys and Tan-I significantly inhibited the expression of INHBA in UUO kidneys (Fig. 4E). Moreover, Tan-I blocked the increased expression of INHBA in AAN kidneys (Fig. 4F).
Tan-I reduces INHBA expression in vitro and in vivo. A The RNA from DMSO or Tan-I treated UUO kidneys was extracted and analyzed by RNA-Seq. A Volcano profiles of RNA sequence between the two groups was shown. B Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis of differential expressed genes between two groups was performed [38,39,40]. C Quantitative PCR analysis of INHBA expression in UUO kidneys treated with DMSO or Tan-I. N = 8–11 in each experimental group and one representative of at least three independent experiments is shown. Data represents mean ± SD. # p < 0.05 vs UUO/DMSO. D NRK-49F cells were starved for 24 h and followed by 24 h treatment with different concentration (0.5 μM, 5 μM, 50 μM) of Tan-I. The expression of inhibin beta-A (INHBA) was analyzed by Western blotting and then quantified. Data represents mean ± SD. #p < 0.05 vs DMSO. ##p < 0.01 vs DMSO. N = 9 in each experimental group and one representative result of at least three independent experiments is shown. E The expression of INHBA in sham or UUO kidneys was analyzed by Western blotting and then quantified. Data represents mean ± SD. *p < 0.05 vs. Sham/DMSO. ##p < 0.01 vs UUO/DMSO. N = 8–11 in each experimental group and one representative of at least three independent experiments is shown. F The expression of INHBA in NS or AAN kidneys was analyzed by Western blotting and then quantified. Data represents mean ± SD. *p < 0.05 vs NS/DMSO. ##p < 0.01 vs AAN/DMSO. N = 8 in each experimental group and one representative of at least three independent experiments is shown. Full-length blots are presented in Supplementary Fig. 4
Tan-I attenuates renal fibrosis through INHBA
INHBA was knocked down by siRNA in NRK-49F cells after 24 h transfection (Fig. 5A). As shown in Fig. 5A, the expression of FN and Snail were down-regulated in INHBA siRNA transfected NRK-49F cells.
Tan-I inhibits fibrotic changes through INHBA. A NRK-49F cells were transfected with nonsense control (NC) or INHBA siRNA (siINHBA). After 24 h, cell lysates were collected and Western blot analysis was performed to measure the expression of INHBA, FN and Snail. Data represent mean ± SD. #p < 0.05 vs NC. ##p < 0.01 vs NC. ###p < 0.001 vs NC. N = 9 in each experimental group and one representative result of at least three independent experiments is shown. B NRF-49F cells were transfected with NC or siINHBA. On the second day, cells were treated 50 μM of Tan-I for another 24 h. Cell lysates were collected and Western blot analysis was performed to measure the expression of INHBA, FN and Snail. Data represents mean ± SD. ##p < 0.01 vs NC/DMSO. NS represents not significant. N = 9 in each experimental group and one representative result of at least three independent experiments is shown. Full-length blots are presented in Supplementary Fig. 5
Figure 5B shows that 50 μM Tan-I markedly reduced the expression of INHBA, FN, and Snail in non-sense control (NC)-siRNA transfected NRK-49F cells. The inhibitory effect of 50 μM Tan-I on the expression of FN and Snail was abolished in INHBA siRNA transfected NRK-49F cells (Fig. 5B).
Discussion
In this study we determined the effect of Tan-I in renal fibrosis. We found that Tan-I dose-dependently inhibited the expression of EMT markers and phosphorylation of Smad3 in rat renal fibroblasts. In two different mouse models, we further showed that Tan-I attenuated renal fibrosis in UUO or AAN kidneys as shown by Masson staining and Western blotting analysis of fibrotic markers. Thus, we conclude that Tan-I inhibits renal fibrosis.
RNA-seq was performed to explore the underlining mechanism of anti-fibrotic effect of Tan-I. 88 differential expressed genes were identified between UUO/DMSO and UUO/Tan-I group, among which two genes (INHBA and BMP7) are involved in TGF-β signaling pathway. The predicted differential expression of BMP7 and INHBA were measured by qPCR, and only the expression of INHBA was significantly different between UUO/DMSO and UUO/Tan-I group. INHBA activates TGF-β signaling pathway and promotes EMT, however its role in renal fibrosis is current unknown [18, 19]. Since the importance and novelty of INHBA study in renal fibrosis, we further verified the expression of INHBA in protein levels. We found that INHBA is upregulated in two fibrotic mouse models, and Tan-I inhibited the expression of INHBA in UUO and AAN kidneys. Moreover, the direct inhibitory effect of Tan-I on INHBA expression was confirmed in vitro. We further showed that knockdown of INHBA reduced the expression of pro-fibrotic markers and abolished the inhibitory effect of Tan-I on the expression of pro-fibrotic markers in renal fibroblasts.
Multiple signaling pathways are regulated by Tan-I, among which nuclear factor erythroid 2 related factor 2 (Nrf2) signaling was identified as an important target of Tan-I [25,26,27]. Nrf2 inhibits inflammation, oxidative stress and protects mitochondria function in chronic kidney diseases [28]. Tan-I can directly activate Nrf2 as its potential agonist or indirectly activate Nrf2 through the Akt signaling pathway [25,26,27]. Thus, whether Tan-I inhibits renal interstitial fibrosis through Nrf2 warrants further study.
The methyltransferase Enhancer of zeste homolog 2 (EZH2) is an important epigenetic regulator and has been intensively studied in the field of renal diseases [29]. EZH2 and its downstream target histone H3 lysine 27 trimethylation (H3K27me3) are upregulated in mouse kidneys with renal interstitial fibrosis [30]. Blockage of EZH2 attenuates renal interstitial fibrosis through inhibition of epithelial-mesenchymal transition (EMT) transition and activation of anti-fibrotic signaling pathways such as PTEN and Smad7 [29,30,31]. A recent study showed that Tan-I is an inhibitor of EZH2, which directly binds to EZH2 and inhibits its enzymatic activity on histone methylation [32]. Thus, EZH2 mediated epigenetic gene regulation could be a mechanism underlying the role of Tan-I in renal fibrosis.
Inflammation is a major event underlying the progression of CKD [33]. The innate immunity pathways nuclear factor ĸB (NF-ĸB) is a pivotal driver of inflammation and activation of NF-ĸB triggers the section of many pro-inflammatory factors [34]. Inhibition of NF-ĸB attenuates renal fibrosis in animal models [35]. The anti-inflammatory effect of Tan-I has been reported recently [36]. Tan-I inhibits IL-1β-induced NF-ĸB activation in chondrocytes, a model for osteoarthritis [37]. Tan-I together with Tanshinone IIA/B attenuate LPS-induced inflammation in mastitis probably through the NF-κB signaling pathway [36]. Whether Tan-I inhibits renal fibrosis through the NF-κB signaling pathway should be investigated in the future.
Conclusion
We conclude that Tan-I attenuates fibrosis in fibrotic kidneys through down-regulation of INHBA. Our study suggests that Tan-I could be a novel and effective approach to treat CKD patients with renal interstitial fibrosis.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- Tan-I:
-
Tanshinone I
- NRK-49F:
-
Rat renal fibroblasts
- UUO:
-
Unilateral ureteral obstructionn
- AAI:
-
Aristolochic acid I
- INHBA:
-
Inhibin beta-A
- CKD:
-
Chronic kidney disease
- ECM:
-
Ext-racellular matrix
- TGF-β:
-
Transforming growth factor beta
- EMT:
-
Epithelial mesenchy-mal transition
- FN:
-
Fibronectin
- α-SM-A:
-
α Smooth muscle actin
- AAN:
-
Aristolochic acid nephropathy
- Col-I:
-
Collagen-I
- pSmad3:
-
Phosphorylated Smad-3
- SD:
-
Standard deviati-on
- KEGG:
-
Kyoto Encyclopedia of Genes and Genome-s
- a-ctivin A:
-
A subunit of inhibin A
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
References
Webster AC, Nagler EV, Morton RL, Masson P. Chronic Kidney Disease. Lancet. 2017;389(10075):1238–52.
Eddy AA. Overview of the cellular and molecular basis of kidney fibrosis. Kidney Int Suppl (2011). 2014;4(1):2–8.
Lee SB, Kalluri R. Mechanistic connection between inflammation and fibrosis. Kidney Int Suppl. 2010;119:S22–6.
Zhou T, Luo M, Cai W, Zhou S, Feng D, Xu C, et al. Runt-Related Transcription Factor 1 (RUNX1) Promotes TGF-beta-Induced Renal Tubular Epithelial-to-Mesenchymal Transition (EMT) and Renal Fibrosis through the PI3K Subunit p110delta. EBioMedicine. 2018;31:217–25.
Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol. 2016;12(6):325–38.
Xu BH, Sheng J, You YK, Huang XR, Ma RCW, Wang Q, et al. Deletion of Smad3 prevents renal fibrosis and inflammation in type 2 diabetic nephropathy. Metabolism. 2020;103:154013.
Inazaki K, Kanamaru Y, Kojima Y, Sueyoshi N, Okumura K, Kaneko K, et al. Smad3 deficiency attenuates renal fibrosis, inflammation, and apoptosis after unilateral ureteral obstruction. Kidney Int. 2004;66(2):597–604.
Grande MT, Sanchez-Laorden B, Lopez-Blau C, De Frutos CA, Boutet A, Arevalo M, et al. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat Med. 2015;21(9):989–97.
Cui S, Chen S, Wu Q, Chen T, Li S. A network pharmacology approach to investigate the anti-inflammatory mechanism of effective ingredients from Salvia miltiorrhiza. Int Immunopharmacol. 2020;81:106040.
Yan Y, Su W, Zeng S, Qian L, Chen X, Wei J, et al. Effect and mechanism of Tanshinone I on the radiosensitivity of lung cancer cells. Mol Pharm. 2018;15(11):4843–53.
Zhou J, Jiang YY, Chen H, Wu YC, Zhang L. Tanshinone I attenuates the malignant biological properties of ovarian cancer by inducing apoptosis and autophagy via the inactivation of PI3K/AKT/mTOR pathway. Cell Prolif. 2020;53(2):e12739.
Wang W, Li J, Ding Z, Li Y, Wang J, Chen S, et al. Tanshinone I inhibits the growth and metastasis of osteosarcoma via suppressing JAK/STAT3 signalling pathway. J Cell Mol Med. 2019;23(9):6454–65.
Kim JY, Kim KM, Nan JX, Zhao YZ, Park PH, Lee SJ, et al. Induction of apoptosis by tanshinone I via cytochrome c release in activated hepatic stellate cells. Pharmacol Toxicol. 2003;92(4):195–200.
Tao S, Zheng Y, Lau A, Jaramillo MC, Chau BT, Lantz RC, et al. Tanshinone I activates the Nrf2-dependent antioxidant response and protects against As(III)-induced lung inflammation in vitro and in vivo. Antioxid Redox Signal. 2013;19(14):1647–61.
Feng C, Xie X, Wu M, Li C, Gao M, Liu M, et al. Tanshinone I protects mice from aristolochic acid I-induced kidney injury by induction of CYP1A. Environ Toxicol Pharmacol. 2013;36(3):850–7.
Okano M, Yamamoto H, Ohkuma H, Kano Y, Kim H, Nishikawa S, et al. Significance of INHBA expression in human colorectal cancer. Oncol Rep. 2013;30(6):2903–8.
Asashima M, Ariizumi T, Malacinski GM. In vitro control of organogenesis and body patterning by activin during early amphibian development. Comp Biochem Physiol B Biochem Mol Biol. 2000;126(2):169–78.
Chen ZL, Qin L, Peng XB, Hu Y, Liu B. INHBA gene silencing inhibits gastric cancer cell migration and invasion by impeding activation of the TGF-beta signaling pathway. J Cell Physiol. 2019;234(10):18065–74.
Yu Y, Wang W, Lu W, Chen W, Shang A. Inhibin beta-A (INHBA) induces epithelial-mesenchymal transition and accelerates the motility of breast cancer cells by activating the TGF-beta signaling pathway. Bioengineered. 2021;12(1):4681–96.
Wu M, Lin P, Li L, Chen D, Yang X, Xu L, et al. Reduced asymmetric dimethylarginine accumulation through inhibition of the type I protein arginine methyltransferases promotes renal fibrosis in obstructed kidneys. FASEB J. 2019;33(6):6948–56.
Xu L, Tan B, Huang D, Yuan M, Li T, Wu M, et al. Remdesivir Inhibits Tubulointerstitial Fibrosis in Obstructed Kidneys. Front Pharmacol. 2021;12:626510.
Wei Y, Gao J, Qin L, Xu Y, Wang D, Shi H, et al. Tanshinone I alleviates insulin resistance in type 2 diabetes mellitus rats through IRS-1 pathway. Biomed Pharmacother. 2017;93:352–8.
Chevalier RL, Forbes MS, Thornhill BA. Ureteral obstruction as a model of renal interstitial fibrosis and obstructive nephropathy. Kidney Int. 2009;75(11):1145–52.
Sellmayr M, Hernandez Petzsche MR, Ma Q, Kruger N, Liapis H, Brink A, et al. Only hyperuricemia with crystalluria, but not asymptomatic hyperuricemia, drives progression of chronic kidney disease. J Am Soc Nephrol. 2020;31(12):2773–92.
Zhuo Y, Yuan R, Chen X, He J, Chen Y, Zhang C, et al. Tanshinone I exerts cardiovascular protective effects in vivo and in vitro through inhibiting necroptosis via Akt/Nrf2 signaling pathway. Chin Med. 2021;16(1):48.
Wu YT, Xie LP, Hua Y, Xu HL, Chen GH, Han X, et al. Tanshinone I Inhibits Oxidative Stress-Induced Cardiomyocyte Injury by Modulating Nrf2 Signaling. Front Pharmacol. 2021;12:644116.
de Oliveira MR, Schuck PF, Bosco SMD. Tanshinone I Induces Mitochondrial Protection through an Nrf2-Dependent Mechanism in Paraquat-TreatedHuman Neuroblastoma SH-SY5Y Cells. Mol Neurobiol. 2017;54(6):4597–608.
Stenvinkel P, Chertow GM, Devarajan P, Levin A, Andreoli SP, Bangalore S, et al. Chronic inflammation in chronic kidney disease progression: role of Nrf2. Kidney Int Rep. 2021;6(7):1775–87.
Li T, Yu C, Zhuang S. Histone Methyltransferase EZH2: a potential therapeutic target for kidney diseases. Front Physiol. 2021;12:640700.
Zhou X, Zang X, Ponnusamy M, Masucci MV, Tolbert E, Gong R, et al. Enhancer of Zeste Homolog 2 Inhibition Attenuates Renal Fibrosis by Maintaining Smad7 and Phosphatase and Tensin Homolog Expression. J Am Soc Nephrol. 2016;27(7):2092–108.
Zhou X, Xiong C, Tolbert E, Zhao TC, Bayliss G, Zhuang S. Targeting histone methyltransferase enhancer of zeste homolog-2 inhibits renal epithelial-mesenchymal transition and attenuates renal fibrosis. FASEB J. 2018;32:fj201800237R.
Huang Y, Yu SH, Zhen WX, Cheng T, Wang D, Lin JB, et al. Tanshinone I, a new EZH2 inhibitor restricts normal and malignant hematopoiesis through upregulation of MMP9 and ABCG2. Theranostics. 2021;11(14):6891–904.
Lv W, Booz GW, Wang Y, Fan F, Roman RJ. Inflammation and renal fibrosis: recent developments on key signaling molecules as potential therapeutic targets. Eur J Pharmacol. 2018;820:65–76.
Chuang PY, Menon MC, He JC. Molecular targets for treatment of kidney fibrosis. J Mol Med (Berl). 2013;91(5):549–59.
Andrade-Oliveira V, Foresto-Neto O, Watanabe IKM, Zatz R, Camara NOS. Inflammation in renal diseases: new and old players. Front Pharmacol. 2019;10:1192.
Yang L, Zhou G, Liu J, Song J, Zhang Z, Huang Q, et al. Tanshinone I and Tanshinone IIA/B attenuate LPS-induced mastitis via regulating the NF-kappaB. Biomed Pharmacother. 2021;137:111353.
Wang X, Fan J, Ding X, Sun Y, Cui Z, Liu W. Tanshinone I inhibits IL-1beta-induced apoptosis, inflammation and extracellular matrix degradation in chondrocytes CHON-001 cells and attenuates murine osteoarthritis. Drug Des Devel Ther. 2019;13:3559–68.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–51.
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545–51.
Acknowledgements
Not applicable.
Funding
This work was supported by Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (PWZxq2017-07), The Three Year Action Plan Project of Shanghai Accelerating Development of Traditional Chinese Medicine (ZY(2018–2020)-CCCX-2003–08) and National Natural Science Foundation of China (81873617) to CY, Scientific Research Foundation of Shanghai Municipal Commission of Health and Family Planning (201740193) to MW.
Author information
Authors and Affiliations
Contributions
MW and CY conceived and coordinated the study. CY founded the project. MW wrote the paper. FY conducted the in vitro experiments. MW, FY and DH performed the animal experiments. FY performed and analyzed the Western blotting. All authors reviewed the results and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The animal procedures were approved by the Animal Experiment Ethics Committee of Shanghai University of Traditional Chinese Medicine in accordance with the principles outlined in the NIH Guide for the Care and Use of Laboratory Animals (PZSHUTCM18111601).
Consent for publication
Not applicable.
Competing interest
The authors have no conflicts of interest to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
The original images of Western blot assay in figure 1
Additional file 2.
The original images of Western blot assay in figure 2
Additional file 3.
The original images of Western blot assay in figure 3
Additional file 4.
The original images of Western blot assay in figure 4
Additional file 5.
The original images of Western blot assay in figure 5
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wu, M., Yang, F., Huang, D. et al. Tanshinone I attenuates fibrosis in fibrotic kidneys through down-regulation of inhibin beta-A. BMC Complement Med Ther 22, 110 (2022). https://doi.org/10.1186/s12906-022-03592-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-022-03592-3