Abstract
Background
Plant extracts were evaluated on poultry bacteria known to be threatening public health. This is to develop better bio-therapeutic agents from plant origin.
Methods
Bacteria were isolated from water, feed, crop, gizzard and faeces of layer chicken. Isolates of interest (Escherichia coli, Salmonella enteritidis, Pseudomonas aeruginosa and Klebsiella oxytoca) were subjected to antibiotic susceptibility test. Resistant strains were further evaluated against different plant extracts in comparison to Meropenem (control) using agar diffusion method.
Results
E. coli had the highest occurrence (53 %), followed by P. aeruginosa (25 %) and then S. enteritidis (13 %) while the least was K. oxytoca (9 %). Virtually all the isolates exhibited multi-antibiotic resistance (MAR) with gross resistance to Amoxicillin, Erythromycin and Cefuroxine. P. aeruginosa (75 %), S. enteritidis (75 %) and E. coli (63 %), had the highest MAR. Out of the 11 (100 %) plant extracts evaluated, 7 (64 %) were outstanding and showed varied levels of antibacterial activity. Specifically, methanol extract of Mangifera indica Julie cultivar leaf (MJLM) had the highest antibacterial activity, followed by Euadenia trifoliata stem bark (TB03) and Euadenia eminens leaf (TB05). P. aeruginosa was highly susceptible (81.81 %) to the extracts, followed by S. enteritidis (63.64 %) and then E. coli (27.27 %).
Conclusions
MJLM and other extracts have proven to be promising extracts in which to search for bioactive components that can be developed into therapeutic drugs. This may help in the management of antibiotic resistant bacterial isolates from poultry chicken threatening public health.
Similar content being viewed by others
Background
Poultry is one of the world’s fastest growing sources of meat, and its share in the world meat production has increased from 15 % to 30 % [1]. The modern production unit can produce market ready broiler chicken in less than six weeks. This development arose from genetic selection, improved feeding and health management practices involving use of antibiotics as therapeutic agents [2, 3]. Antibiotics have improved poultry performance effectively and economically [3, 4] but an increase in numbers of antibiotic-resistant bacterial strains [5] such as E. coli [6–8], Salmonella spp. [9] and Pseudomonas spp. [10] continue to occur [4, 11].
The high incidence and rising frequency of antibiotic resistance among the bacteria populating poultry chicken presents a public health hazard [5]. These antibiotic resistant bacteria can be transmitted from poultry to humans through the food chain with serious consequences on public health [5, 12]. This, therefore, necessitates a need for better antimicrobial drugs. Plants have been documented as one of the sources that possesses antimicrobial traits which are chiefly synthesized during secondary metabolism [13–15]. Plant based antimicrobial compounds have great therapeutic potentials as they can serve the purpose without any side effects associated with synthetic drugs [16]. The inherent utility and practical applications of plant extracts such as garlic, cinnamon, tulsi, ginger, turmeric, lemon, neem, yucca, thyme and rosemary have been explored for improving poultry health as well as production with fruitful results [17, 18]. Though, it has been reported that some plant based chemotherapeutic agents may be ineffective on emerging resistant bacterial strains [19, 20], therefore, further work still needs to be done to search for more effective plant based chemotherapeutic agents especially for poultry chicken management.
Generally, plants contain bioactive components [21, 22], that are less toxic and environmentally friendly [23]. Their antibacterial activity have been documented against methicillin-resistant Staphylococcus aureus and a variety of other bacteria [24, 25] such as Escherichia coli, Klebsiella pneumonia [26], Pseudomonas aeruginosa, Proteus mirabilis [27], Brevibacterium ammoniagenes, Streptococcus mutans and Propionibacterium acnes [28]. Despite the fact that global interest has been shifted to plant based antimicrobials [29], many plants still remain largely untapped against antibiotic resistant bacterial isolates. This study therefore investigated not only susceptibility of isolated bacteria to different antibiotics but also antibacterial activity (in vitro) of plant extracts on selected antibiotic resistant bacterial isolates. These isolates were obtained from water, feed, crop, gizzard, and faeces of poultry layer chicken. The isolates were characterized and assayed as multi-antibiotic resistant bacteria and then subjected to different plant extracts. Apart from Mangifera indica (Mango), other plant extracts used have not previously been evaluated on the antibiotic resistant bacterial isolates from poultry chicken.
Methods
Collection of samples, bacterial isolation and identification
Poultry practicing rural community, Ilora in Oyo State, Southwestern Nigeria was used as our collection site. The following samples, namely; water, feed, crop and gizzard contents and faecal materials were randomly collected in 3 replicates per sample from at least 5 different poultry farms that were about 100 km apart. Water samples were aseptically collected from nipples, reservoirs and storage tanks of layer chickens. Feed samples were obtained from layer chicken feed while the crop and gizzard were obtained from already sacrificed layer chickens. Fresh faecal materials were also collected from droppings of layer chickens kept in battery cage system. After collection, samples were aseptically transported to laboratory for specific bacterial contents. Apart from water samples, stomacher (Seward Stomacher*80 Lab System, England) at 60 s normal speed was used to aid maceration of solid samples in stomacher bag (1 g in 9 ml of sterile water) for successful serial dilution–pour plate isolation [30] of bacteria. Inoculated nutrient agar (LabM Limited, Lancashire, United Kingdom) and MacConkey agar (Medical Market Limited, United Kingdom) plates were incubated at 37 ± 2 °C for 24–48 h [31]. Distinct colonies were randomly selected and were streaked onto new plates until pure cultures were obtained. The bacterial isolates were identified based on morphological and biochemical tests. Gram and endospore staining, catalase, oxidase, indole and citrate test were carried out following standard protocols [32]. Utilisation of lactose, sucrose, glucose and gas production was confirmed using triple sugar iron (TSI; LabM Limited, Lancashire, United Kingdom) agar slant [33]. Motility test was also carried out by stabbing sulphide indole motility (SIM; Rapid Lab. Colchester Essex, United Kingdom) agar vertically with each bacterium isolate and appearance of turbidity confirmed motility after incubation for 24–48 h [33]. The overall morphological and biochemical characteristics were subjected to identification using Bergey’s Manual of Determinative Bacteriology [34].
Antibiotic resistance profile of test strains
Prior to antibiotic sensitivity test, McFarland standard corresponding to 0.5 was prepared [35, 36]. Turbidity was confirmed to have optical density (OD) of 0.08–0.10 at 625 nm using photo-electric colorimeter (Callen Komp, England). The antibiotics susceptibility of the isolates was performed using Gram specific antibiotics (Rapid Labs, United Kingdom) which include: Ceftazidime (30 μg), Ciprofloxacin (5 μg), Ofloxacin (5 μg), Amoxicillin (30 μg), Ampicillin (10 μg), Nitrofurantoin (300 μg), Gentamicin (10 μg), Cefuroxime (30 μg), Cloxacillin (5 μg), Erythromycin (10 μg) and Ceftriaxone (30 μg). Using a sterile inoculating loop, the distinct colony of the isolates was emulsified in 3–4 ml of sterile physiological saline and turbidity of the bacterial suspension was matched to the turbidity of the standard. The bacterial suspension was swabbed with sterile cotton swab evenly on Mueller Hinton (MH; Cypress Diagnostic, Belgium) agar in Petri - dishes by rotating plate at approximately 60°. The multi - antibiotic discs (Rapid Labs, United Kingdom) was placed aseptically onto inoculated three replicated plates with respect to each isolate using sterile forceps and incubated aerobically at 37 ± 2 °C for 24–48 h [32, 37]. Diameter of the zones of inhibition were measured with a ruler and recorded in millimeter (mm). The interpretation of the results was done using interpretative chart according to CLSI [38]. Based on the interpretation, the bacterial isolates were classified either as susceptible or resistant.
Plant preparation and extraction
The leaves and stem bark of three cultivars (Julie, Edward, and Ogbomosho) of Mangifera indica were collected from cultivated Mango orchards at the National Horticultural Research Institute (NIHORT), Ibadan in Nigeria and were authenticated by the scientist in charge. It was then identified at the herbarium unit of the Department of Botany, University of Ibadan, Nigeria. The herbarium number were as follows: Julie cultivar - UIH 22514J, Edward cultivar - UIH 22514E, and Ogbomoso cultivar - UIH 22514O. The stem-bark of Erythrophleum suaveolens (ID: IFH-17542) as well as the leaves and stem-bark of Euadenia eminens (ID: IFH-16540) and Euadenia trifoliata (ID: IFH-16539) were collected at a farmland in Lalupon village, Oyo State, Nigeria and were confirmed at herbarium unit, Department of Botany, Obafemi Awolowo University, Ife, Nigeria. The milled, air dried plant samples were extracted with methanol. The extracts obtained were filtered and concentrated under vacuum to give the respective crude extracts. The following codes were given to the extracts: Erythrophleum suaveolens leaves (TB01), Euadenia trifoliata leaves (TB02), Euadenia trifoliata stem-bark (TB03), Euadenia eminens stem-bark (TB04), Euadenia eminens leaves (TB05), methanol extract of Mango Juliet cultivar leaves (MJLM), methanol extract of Mango Juliet cultivar stem-bark (MJSBM), methanol extract of Mango Ogbomoso cultivar stem-bark (MOSBM), methanol extract of Mango Edward cultivar stem-bark (MESBM) and methanol extract of Mango Edward cultivar leaves (MELM).
Antimicrobial activity of plant extracts
In preparation for the antimicrobial assay, 50 ml of DMSO (Dimethyl sulfoxide, Sigma-Aldrich) was added to 1 g of each extract in sterile vial bottles to give a stock solution of 1 g/50 ml (w/v) concentration and kept at 4 °C until use. Antibiotic disc diffusion assay [37] with slight modifications [39, 40] was used. Briefly, 24 h old bacterial isolates were standardised against 0.5 McFarland standard [35, 36]. The sterile swab stick was used to pick inoculum from already standardised broth culture in peptone water, and was evenly spread on MH (Cypress Diagnostic, Belgium) agar plates. Sterilised 8 mm cork borer was used to make a hole in the three replicated agar plates and filled with 0.1 ml of each extract (1 g/50 ml equivalent to 0.02 g/ml) while 0.1 ml of 0.02 g/ml of Meropenem (Ranbaxy Laboratories limited, India) was used as the control. The plates were incubated at 37 ± 2 °C for 24–48 h. The diameter of the zone of inhibition around each hole was measured in millimeters (mm) and the mean value was calculated.
Quantitative evaluation of antimicrobial activity
Percentage activity: This demonstrates the total antimicrobial potency of a particular extract. It shows number of bacteria found susceptible to one particular extract [41].
Bacterial susceptibility index (%) (BSI): This is used to compare the relative susceptibility between bacterial isolates. The BSI values range from 0% (resistant to all samples) to 100% (susceptible to all samples) [41].
Statistical analyses
Treatments were compared using SAS software, version 9.1 (SAS Institute, Cary, NC, USA) [42]. From in vitro antibiotic susceptibility assay, dependent variables were subjected to analysis of variance (ANOVA), followed by post hoc pairwise comparisons using the Student-Newman-Keuls multiple-range test. Three out of the four isolates with high MAR were subjected to eleven extracts in an in vitro experiment. Values obtained from the effectiveness of plant extracts on the isolates were also compared by ANOVA and Student-Newman-Keuls test.
Results
Characteristics and occurrence of bacterial isolates
A total of thirty-two Gram negative bacteria were isolated from different poultry chicken sources. The isolates exhibited morphological characteristics that ranges from green, light yellow, deep yellow, rough surface and opaque to circular (Table 1). Bacterial isolates were identified based on morphological characteristics and biochemical test with key interest in P. aeruginosa, S. enteritidis, E. coli and K. oxytoca (Table 2). P. aeruginosa and K. oxytoca were predominantly isolated from water used in poultry chicken management. E. coli was isolated from crop, gizzard and poultry chicken faeces while only S. enteritidis originated from chicken faeces. Irrespective of the sources, E. coli dominated, followed by P. aeruginosa while S. enteritidis and K. oxytoca were the least.
Antimicrobial sensitivity assay
The phenotypic antibiotic resistance profile of the Gram negative bacteria isolated from poultry chicken sources was studied using 50 % as the breaking point for the percentage effectiveness. There was 87–100 % resistance to Cefuroxine across the bacteria except S. enteritidis that showed no resistance, however, 75–100 % resistance to Amoxicillin. All the isolates showed gross resistance to Amoxicillin, Erythromycin and Cefuroxine. Surprisingly, K. oxytoca showed outstanding sensitivity to Ceftazidine and Cloxacillin in comparison to other isolates that were resistant to them. P. aeruginosa and S. enteritidis displayed resistance to Gentamycin and they both similarly had the highest MAR (75 %), followed by E. coli (63 %) (Table 3).
Antibacterial activity of the plant extracts
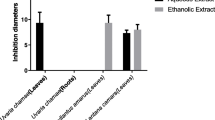
Bacterial isolates (P. aeruginosa, S. enteriditis and E. coli) that showed high MAR were selected and subjected to different plant extracts. Out of the 11 extracts evaluated, 7 (64 %) were effective and vary in their antibacterial activities against MAR isolates (Table 3). Based on percentage activity, MJLM had the highest antibacterial activity (100 %), followed by TB02, TB03, TB05, MELM, MESBM and MOSBM (66.7 %) while the least were TB01, TB04, MOLM and MJSBM (33.33 %) (Fig. 1). MJLM exhibited broad spectrum inhibitory activity and also compared favourably with Meropenem (control) against the selected MARS bacterial isolates (Table 3; Fig. 2). Specifically, TB01, TB02, TB03, TB05, MJLM, MELM, MESBM, MOSBM and MJSBM were effective on Pseudomonas spp. TBO3, MESBM, MOSBM and MOLM demonstrated partial inhibitory activity on S. enteritidis in comparison to E. coli that exhibited resistance to virtually more than 60 % of the plant extracts (Table 4; Fig. 3).
Discussion
Interest in poultry bacteria specifically those that threaten public health motivated this study. Based on phenotypic antibiotic susceptibility test, the level of resistance exhibited by the test isolates on selected antibiotics is alarming. This is an indication that indiscriminate use of conventional antimicrobials has led to a steady increase in the antibiotic resistance and the low-income countries which are home to the majority of the world’s population are particularly affected by this phenomenon [14]. With the exception of K. oxytoca, the antibacterial activity of plant extracts was centred on E. coli, S. enteritidis and P. aeruginosa because of their high MAR.
Apart from the occurrence of E. coli in the crop, gizzard and faeces of poultry chickens, their resistance to antibiotics is a great concern. This shows that preventive medications are still given to chicken in order to reduce mortality [43]. The application of antibiotics will not only increase the resistance in pathogenic strains but can also lead to resistance in the endogenous flora of humans and animals [44]. Following the consumption of poultry meat specifically chicken, MAR bacterial strains may spread to the human population, which can lead to the transfer of genes coding for resistance [45]. Our observation shows that the resistance rate of E. coli to Amoxicillin, Erythromycin, Cefuroxine, Ceftazidine and Cloxacillin presents a serious cause for concern considering the fact that the uninformed farmers may continue to use increasing level of ineffective antibiotics in the management of infection in farms with possible residues in poultry meat, eggs and other products meant for human consumption [46].
One of the ways to handle antibiotic resistant E. coli is to develop new and novel antimicrobial agents specifically from plant origin. From this study, the methanol extract of Mangifera indica Julie cultivar leaves demonstrated antibacterial effectiveness on antibiotic resistant E. coli [47]. This is an indication that the leaves of Mangifera indica contain inhibitory substances active against E. coli [48] in comparison to Euadenia trifoliata that was ineffective. This observation is similar to that of Euadenia eminens root extract evaluated against E. coli by Dickson et al. [49] but contrary to that of Amole et al. [50]. The resistance of E. coli to the extracts [51] is similar to what was obtained for Amoxicillin, Erythromycin, Cefuroxine, Ceftazidine and Cloxacillin. Although, in this case, the crude extract used may not be active enough as potency of each phytoactive components may have been affected by each other. This suggests that further work still needs to be done to evaluate the specific phytoactive compound present in all the tested plant extracts so as to ascertain their efficacy on this antibiotic resistant E. coli.
S. enteritidis occurrences in faeces of poultry chicken may be as a result of improperly disposed poultry wastes [52]. With reference to Nigeria, wastes from commercial poultry farms are not properly disposed and most rural farmers use these wastes as manure, which are often kept at the backyards before moving them to the farm. These poultry wastes may serve as source of enteric organisms that habour novel resistance factors for birds, including chicken that feed on such wastes. The isolated S. enteritidis was generally resistant to Gentamycin, Amoxicillin, Cloxacillin, Erythromycin, Cefuroxime and Ceftazidine. The observed high resistance against these antibiotics probably reflects the high usage of the drugs in the study sites. This could be because these antibiotics are readily available and farmers see them as first point of contact broad-spectrum to treat their chickens [53, 54]. As a result of this, these drugs may have become seriously compromised and probably are currently ineffective.
The high rates of antibiotic resistance in S. enteritidis may be more difficult to treat with synthetic antibiotics alone, but rather needs the touch of nature such as bioactive components in plants. Evaluated plant extracts inhibited the growth of S. enteritidis in vitro. Extracts from Euadenia trifoliata leaves and Euadenia trifoliata stem-bark demonstrated antibacterial activity against S. enteritidis. This corroborated the work of Amole et al. [50] using petroleum ether, ethyl acetate and methanol fractions of Euadenia trifoliata leaves against S. typhi. Osundiya et al. [55] also reported similar observation using root extracts of Burkea africana and Combretum adenogonium against S. typhi. The antibacterial activity of methanol extract of Mangifera indica - Julie cultivar leaves on S. enteritidis is in agreement with the work of De and Pal [48] who also used aqueous young leaves extract of Mangifera indica against S. typhi. This suggests that Euadenia trifoliate and Mangifera indica possesses antibacterial compounds which are active against Salmonella species.
The occurrence of P. aeruginosa in feed [56] and water [10, 57] is not new however, our work re-established it. P. aeruginosa exhibited resistance to Erythromycin, Amoxicillin, Cloxacillin, Cefuroxime, Ceftazidine and Gentamycin which corroborated with the work of Kibret and Abera [58]. Pseudomonas species are naturally resistant to many antibiotics due to the permeability barriers afforded by its outer membrane composed of lipopolysaccharide [27]. Resistance of P. aeruginosa to Gentamycin, is unique because it has been documented that P. aeruginosa used to be resistant to certain antibiotics [59] but not Gentamycin, however this may be peculiar to our study site. This suggests that poultry chicken farmers (especially the untrained) in the study site may have indirectly abused Gentamycin as a broad spectrum antibiotic [53, 54] to treat poultry chicken diseases.
P. aeruginosa was resistant to more than 70 % of the antibiotics, but sensitive to more than 80 % of the extracts evaluated. This shows that one of the methods to reduce the resistance of P. aeruginosa to synthetic antibiotics is by using antibiotic resistant inhibitors from plant origin [60, 61]. The effectiveness of the plant extracts is in agreement with Abid et al. [62] who also emphasized that the most sensitive bacterium to tested plants is Pseudomonas. In this study, activity of Euadenia trifoliata [50], Euadenia eminens [49] and Mangifera indica was evident against P. aeruginosa [62]. Interestingly, methanol extracts of Mangifera indica (MJLM) showed more activity against P. aeruginosa [47] in comparison to other extracts. This suggests that Mangifera indica possesses potent and specific inhibitory substance (s) against P. aeruginosa.
Conclusions
Isolated bacteria were resistant to more than two antibiotics. Apart from gross resistance to Amoxicillin, Erythromycin and Cefuroxine, susceptibility to Ceftriaxone was observed across the bacteria investigated. The in vitro study revealed that 70 % of extracts exhibited antibacterial activity against test isolates. MJLM and other extracts have proven to be promising extracts in which to search for bioactive compounds that can be developed into therapeutic drugs. These, therefore, substantiate their therapeutic use in the control of antibiotic resistant P. aeruginosa, S. enteriditis and E. coli threatening public health through poultry chicken.
References
FAO. 2006a: World agriculture: towards 2030/2050 interim report. Rome
Murugkar HV, Rahman H, Kumar A, Bhattacharyya D. Isolation, phage typing and antibiogram of Salmonella from man and animals in Northern India. Indian J Med Res. 2005;122:237–42.
Dhama K, Latheef SK, Mani S, Samad HA, Karthik K, Tiwari R, Khan RU, Alagawany M, Farag MR, Alam GM, Laudadio V, Tufarelli V. Multiple beneficial applications and modes of action of herbs in poultry health and production-a review. Int J of Pharmacol. 2015;11:152–76.
Scanes CG. The global importance of poultry. Poult Sci. 2007;86:1057–8.
Apata DF. Antibiotic resistance in poultry. Int J Poult Sci. 2009;8:226–30.
Yang H, Chen S, White DG, et al. Characterization of multiple-antimicrobial-resistant Escherichia coli isolates from diseased chickens and swine in China. J Clin Microbiol. 2004;42:3483–9.
Miles TD, McLaughlin W, Brown PD. Antimicrobial resistance of Escherichia coli isolates from broiler chickens and humans. BMC Vet Res. 2006;2:1–9.
Talebiyan R, Kheradmand M, Khamesipour F, Rabiee-Faradonbeh M. Multiple antimicrobial resistances of Escherichia coli isolated from chickens in Iran. Vet Med Int. 2014;2014:491418.
Salehi TZ, Mahzounieh M, Saeedzadeh A. The isolation of antibiotic-resistant salmonella from intestine and liver of poultry in shiraz province of Iran. Int J Poult Sci. 2005;4:320–2.
Kebede F. Pseudomonas infection in chickens. J Vet Med Ani Health. 2010;2:55–8.
Byarugaba DK. The structure and importance of the commercial and village based poultry systems in Uganda. Rome: FAO-Consultancy Report, Food and Agriculture Organization of the United Nations; 2007.
McDermott PF, Zhao S, Wagner DD, Simjee S, Walker RD, White DG. The food safety perspective of antibiotic resistance. Anim Biotechnol. 2002;13:71–84.
Kokoska L, Polesny Z, Rada V, Nepovim A, Vanek T. Screening of some Siberian medicinal plants for antimicrobial activity. J Ethnopharmacol. 2002;82:51–3.
Radyowijati A, Haak H. Improving antibiotic use in low-income countries: An overview of evidence on determinants. Soc Sci Med. 2003;57:733–44.
Rusenova N, Parvanov P. Antimicrobial activities of twelve essential oils against microorganisms of veterinary importance. Trakia J Sci. 2009;7:37–43.
El-Mahmood AM, Doughari JH. Phytochemical Screening and Antibacterial evaluation of the leaf and root extracts of Cassia alata Linn. Afr J Pharm Pharmacol. 2008;2:124–9.
Sudarshan S, Fairoze N, Ruban SW, Badhe SR, Raghunath BV. Effect of aqueous extract and essential oils of ginger and garlic as decontaminat in chicken meat. Res J Poult Sci. 2010;3:58–61.
Umashanker M, Shruti S. Traditional Indian herbal medicine used as antipyretic, antiulcer, anti-diabetic and anticancer: A review. Int J Res Pharm Chem. 2011;1:1152–9.
Anon. Antimicrobial resistance: Implications for the food system. Compr Rev Food Sci Food Saf. 2006;5:71–137.
Chastre J. Evolving problems with resistant pathogens. Clin Microbiol Infect. 2008;14 Suppl 3:3–14.
Panda SK, Thatoi HN, Dutta SK. Antibacterial activity and phytochemical screening of leaf and bark extracts of Vitex negundo I. from Similipal biosphere reserve, Orissa. J Med Plant Res. 2009;3:294–300.
Ntonifor NN. Potentials of tropical African spices as sources of reduced-risk pesticides. J Entomol. 2011;8:16–26.
Opara EU, Wokocha RC. Efficacy of some plant extracts on the in vitro and in vivo control of Xanthomonas campestris pv. vesicatoria. Agric J. 2008;3:163–70.
Beg AZ, Ahmad I. Effect of Plumbago zeylanica extract and certain curing agents on multi drug resistant bacteria of clinical origin. World J Microbiol Biotechnol. 2000;16:01–4.
Aqil F, Khan MSA, Owais M, Ahmad I. Effect of certain bioactive plant extracts on clinical isolates of β-lactamase producing methicillin resistant Staphylococcus aureus. J Basic Microbiol. 2005;45:106–14.
Djeussi DE, AK Noumedem J, Seukep JA, Fankam AG, Voukeng IK, Tankeo SB, Nkuete AHL, Kuete V. Antibacterial activities of selected edible plants extracts against multidrug-resistant Gram-negative bacteria. BMC Complement Altern Med. 2013;13:164–72.
Panghal M, Kaushal V, Yadav JP. In vitro antimicrobial activity of ten medicinal plants against clinical isolates of oral cancer cases. Annals of Clin Microbiol Antimicrob. 2011;10:21–7.
Muroi H, Kubo I. Antibacterial activity of anacardic acids and totarol, alone and in combination with methicillin, against methicillin-resistant Staphylococcus aureus. J Appl Bacteriol. 1996;80:387–94.
Abiala MA, Ogunjobi AA, Odebode AC, Ayodele MA. Evaluation of plant extracts as an antagonist to mycelial growth of Mycosphaerella fijiensis morelet. Arch Phytopathol Plant Prot. 2011;44:1711–8.
Reynolds J. Serial dilution protocols. ASM MicrobeLibrary. 30 Sept 2005.http://www.microbelibrary.org/library/laboratory-test/2884-serial-dilution-protocols. Accessed 30 June 2014.
Adeleke MA, Olaitan JO, Abiona O, Canice J, Olajide S, Oluogun W, Fowora M, Okesina A. Molecular characterization and antibiotic susceptibility patterns of bacteria isolated from wara (West African cheese) sold in Osun state, Nigeria. Innov Rom Food Biotechnol. 2014;15:23–30.
Harrigan WF, McCance ME. Laboratory methods in microbiology. New York: Academic; 1996.
Cheesborough M. Biochemical Test to Idenify Bacteria in Laboratory Practice in Tropical Countries. Cheesbrough M. (ed). Cambridge Edition 2006, 2: 63-87
Bergey DH, John GH: Bergeys’s manual of Determinative Bacteriology, 9th ed., Williams and Wilkins, 1994, 4: 181–186
Farland McJ: J Am Medical Assn 1987, 49: 1176–1178.
Chapin KC, Lauderdale T. Reagents, stains, and media: bacteriology. In: Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH, editors. Manual of Clin Microbiol. 8th ed. Washington: ASM Press; 2003. p. 358.
Bauer AW, Kirby W, Sherris J, Turck M. Antibiotic susceptibility testing by a standardized disc method. Am J Clin Pathol. 1966;45:493–6.
Clinical and Laboratory Standards Institute (CLSI): Performance Standards for Antimicrobial Susceptibility Testing; Twenty-fourth Informational Supplement. CLSI document M100-S24, PA 2014, 34: 47–135
Ho PL, Chow KH, Yuen KY, Ng WS, Chau PY. Comparison of a novel, inhibitor-potentiated disc-diffusion test with other methods for the detection of extended-spectrum β-lactamases in Escherichia coli and Klebsiella pneumoniae. J Antimicrob Chemother. 1998;42:49–54.
NCCLS. Performance Standards for Antimicrobial Susceptibility Testing, 14th Informational Supplement Document M2-A8. 8th ed. Wayne: National Committee of Clinical Laboratory Standards; 2004.
Bonjar GHS. Approaches in screening for antibacterial in plants. Asian J of Plant Sci. 2004;31:55–60.
SAS (Statistical Analysis System): SAS/STAT guide for personal computers, version 9.2. SAS Institute Incorporated, Cary, NC. 2009.
Dan SD, Tăbăran A, Mihaiu L, Mihaiu M. Antibiotic susceptibility and prevalence of foodborne pathogens in poultry meat in Romania. J Infect Dev Ctries. 2015;9:035–41. doi:10.3855/jidc.4958.
Kolar M, Antucek P, Ardon RB, Agnerova JV, Ypovska IT, Oskar HD, Alka JVI. Occurrence of antibiotic-resistant bacterial strains isolated in poultry. Vet Med Czech. 2002;47:52–9.
Bogaard AE, Stobberingh EE. Epidemiology of resistance to antibiotics. Links between animals and humans. Int J Antimicrob Agents. 2000;14:327–35.
Anyanwu AL, Fasina FO, Ajayi OT, Rapu I, Fasina MM. Antibiotic resistant Salmonella and Escherichia coli isolated from day-old chicks, vom, Nigeria. Afr J Clin Experim Microbiol. 2010;11:129–36.
Doughari JH, Manzara S. In Vitro Antibacterial Activity of Crude Leaf Extracts of Mangifera indica Linn. Afr J Microbiol Res. 2008;2:67–72.
De PK, Pal A. Effects of aqueous young leaves extract of Mangifera indica on Gram negative microorganisms causing gastro-intestinal disorders. Asian J of Plant Sci Res. 2014;4:23–7.
Dickson RA, Fleischer TC, Ekuadzi E, Komlaga G. Anti-inflammatory, antioxidant, and selective antibacterial effects of Euadenia eminens root bark. Afr J Tradit Complement Altern Med. 2012;2012(9):271–6.
Amole LK, Oyewale AO, Aliyu AB, Garba S. Phytochemical, Cytotoxicity, Antimicrobial and Antioxidant screening of Fractions obtained from Euadenia trifoliata (Capparaceae). Arch Appl Sci Res. 2015;7:35–43.
Yagoup SO, Shami EA, Ahmed B, Asha ZE: Antimicrobial activity of some medicinal plants against some Gram positive, Gram negative and fungi. 2007. Department of Molecular Biology, EL Neelain University, Sudan. www.astf.net Accessed on 20 Aug 2009.
Okoli IC. Antimicrobial resistance profiles of E. coli isolated from free range chickens in urban and rural environments of Imo State, Nigeria. Online J Health Allied Sci. 2006;5:1–6.
Okeke IN, Fayinka ST, Laminkanra A. Antibiotic resistance in E. coli from Nigerian students, 1986–1998. Emerging Infect Dis. 2000;6:393–6.
Chah KF, Okafor SC, Oboegbulem SI. Antimicrobial resistance of none clinical E. coli strains from chickens in Nsukka, South Eastern Nigeria. Nig J Animal Prod. 2003;30:101–6.
Osundiya OO, Oladele RO, Oduyebo OO. Multiple antibiotic resistance (mar) indices of Pseudomonas and Klebsiella species isolates in Lagos university teaching hospital. Afr J Clin Exper Microbiol. 2013;14:164–8. http://dx.doi.org/10.4314/ajcem.v14i3.8.
Okonkwo IO, Nkang AO, Eyerefe OD, Abubakar MJ, Ojezele MO, Amusan TA. Incidence of MultiDrug Resistant (MDR) Organisms in some Poultry Feeds sold in Calabar Metropolis, Nigeria. Brit J Pharmacol Toxicol. 2010;1:15–28.
Adesoji AT, Ogunjobi AA, Olatoye IO. Molecular characterization of selectedmultidrug resistant Pseudomonas from water distribution systems in southwestern Nigeria. BMC Ann Clin Microbiol Antimicrob. 2015;14:39–50.
Kibret M, Abera B. Antimicrobial susceptibility patterns of E. coli from clinical sources in northeast Ethiopia. Afr Health Sci. 2011;11 Suppl 1:S40–5.
Kelley TR, Pancorbo OC, Merka WC, Barnhart HM. Antibiotic resistance of bacterial litter isolates. Poult Sci. 1998;77:243–7.
Iinuma M, Tsuchiya H, Sato M, Yokoyama J, Ohyama M, Ohkawa Y, Tanaka T, Fujiwara S, Fujii T. Flavanones with potent antibacterial activity against methicillin-resistant Staphylococcus aureus. J Pharm Pharmacol. 1994;46:892–5.
Kim H, Park SW, Park JM, Moon KH, Lee CK. Screening and isolation of antibiotic resistant inhibitors from herb materials I - Resistant Inhibition of 21 Korean Plants. Nat Prod Sci. 1995;1:50–4.
Abid AJ, Almashta SA, Tolaifih ZA. Antibacterial activity of some plant extracts against some bacterial species. Med J Babylon. 2014;5:1–4.
Acknowledgements
The authors hereby acknowledge the technical support of Okunlola Idowu, Akande Akinwumi, Olusola Olumide, Akanji Abolade, Dibiagwu Theresa and Ologun Olusanya. We also appreciate the Poultry Association of Nigeria (PAN), Ilora chapter for their cooperation.
Funding
No funding was obtained for this study.
Availability of data and materials
All the data supporting these findings is contained within the manuscript.
Authors’ contribution
MA and JO designed and performed the experiment. MA wrote the manuscript with significant contribution from JO. OB, OA and SA extracted the crude bioactive components from the plants. Reagents and chemicals were supplied by all the authors, and as well read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The concept of the research was properly explained to all the Poultry (Chicken) farmers of the Poultry Association of Nigeria (PAN), Ilora Chapter, Oyo State. Nigeria. Their consents were sought by administration of Informed Consent Form (ICF) and were given the liberty to give their consent without coercion. The risk of collecting crop and gizzard from their sacrificed hens was explained in term of cost value of the gizzard and they were given the freedom to decide. Also, the benefit of the research was duly explained to them like the search for novel anti-microbial agents. None of the farmers was placed under coercion. Samples were coded so as to ensure protection of privacy. Moreso, since the design concept of this study revealed that the sources of sample do not affect humans and animals. The only sample obtained is from crop and gizzard of chicken that were sacrificed by the poultry farmers. In addition, in – case of economy loss, the farmer had given their consent. Therefore, the ethical committee of Faculty of Natural Sciences, Ajayi Crowther University, Oyo State, Nigeria (FNS/ERD/201600003) waived the need for ethical approval in this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Abiala, M., Olayiwola, J., Babatunde, O. et al. Evaluation of therapeutic potentials of plant extracts against poultry bacteria threatening public health. BMC Complement Altern Med 16, 417 (2016). https://doi.org/10.1186/s12906-016-1399-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-016-1399-z