Abstract
Background
Gamisoyo-san (GMSYS) is a traditional herbal formula used to treat insomnia, dysmenorrhea, and infertility in Korea. The purpose of this study was to investigate the anti-inflammatory effect and action mechanisms of GMSYS in lipopolysaccharide (LPS)-stimulated RAW 264.7 macrophages.
Methods
The anti-inflammatory effects of GMSYS were investigated using nitric oxide (NO) assay and ELISAs for prostaglandin E2 (PGE2), tumor necrosis factor-α (TNF-α), and interleukin-6 (IL-6). The anti-inflammatory action mechanisms of GMSYS were evaluated using Western blotting for inducible nitric oxide synthase (iNOS), cyclooxygenase-2 (COX-2), and activation of nuclear transcription factor kappa B (NF-κB) and mitogen-activated protein kinases (MAPKs).
Results
GMSYS significantly inhibited the LPS-induced production of NO, PGE2, TNF-α, and IL-6 compared with the vehicle-treated cells. GMSYS consistently downregulated the expression of iNOS and COX-2 mRNA induced by LPS. In addition, pretreatment with GMSYS suppressed the LPS-induced activation of NF-κB and MAPKs such as p38, extracellular signal-regulated kinase (ERK), and c-Jun N-terminal kinase (JNK).
Conclusions
Our results indicate that the anti-inflammatory effects of GMSYS in RAW 264.7 macrophages are associated with inhibition of the release of inflammatory mediators and cytokines through the suppression of MAPK and NF-κB activation. These findings suggest that GMSYS may be a useful therapeutic candidate for the prevention or treatment of inflammatory diseases.
Similar content being viewed by others
Background
Inflammation is a complex process regulated by a cascade of inflammatory mediators, including nitric oxide (NO), prostaglandin E2 (PGE2), and cytokines, including tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6), that are produced by activated macrophages [1, 2].
NO is generated from l-arginine by nitric oxide synthase (NOS) and plays an important role in the regulation of physiological responses [3]. Among the isoforms of NOS, the expression of inducible NOS (iNOS) is specifically stimulated by cytokines and bacterial lipopolysaccharide (LPS) [4]. Prostaglandins are key inflammatory mediators that are produced from arachidonic acid by cyclooxygenase (COX). Among the isoforms of COX, COX-2 is induced after exposure to specific stimulants including cytokines and LPS. The induction of COX-2 produces a large amount of PGE2 that causes inflammatory reactions [5]. These inflammatory mediators and cytokines are critically associated with pain, fever, edema, and recruitment of additional immune cells to the site of inflammation [6]. However, overproduction of inflammatory mediators and cytokines is associated with tissue damage [7]. Therefore, inhibiting the release of inflammatory mediators and cytokines could be beneficial in attenuating the damage caused by inflammatory diseases.
Mitogen-activated protein kinases (MAPKs) such as p38, extracellular signal-regulated kinase (ERK), and c-Jun N-terminal kinase (JNK) regulate inflammatory and immune responses, and their signaling pathways are involved in LPS-induced iNOS and COX-2 expression in macrophages [8, 9]. In addition, nuclear transcription factor kappa B (NF-κB) is crucial during inflammation [10]. The regulatory free NF-κB translocates to the nucleus during inflammation, where it binds to κB-binding sites in the promotor regions of target genes and induces the transcription of inflammatory mediators and cytokines such as iNOS, COX-2, TNF-α, and IL-6 [11]. Thus, there may be an opportunity to improve the development of the inflammatory response by regulating the activation of the MAPK and NF-κB pathways.
In Korea, Gamisoyo-san (GMSYS) has been widely used for the treatment of dysmenorrhea, insomnia, and anxiety [12]. GMSYS is effective for treatment of sleep disturbances, headache, dizziness in postmenopausal women, depressive symptoms in premenstrual dysphoric disorder, and for tardive dyskinesia related to the use of antipsychotic drugs [13–15]. In addition, GMSYS has been reported to exert antistress, antidepressive, and antioxidant activities [12, 16–20]. Despite previous studies, there have been no reports on the effects and molecular mechanisms of GMSYS on inflammatory responses.
In the present study, we investigated the effect of GMSYS on the expression and release of inflammatory mediators and cytokines including NO, iNOS, PGE2, COX-2, TNF-α, and IL-6 from LPS-stimulated RAW 264.7 macrophages. In addition, we studied the molecular mechanisms possibly involved in the regulation of inflammatory responses by GMSYS.
Methods
Preparation of GMSYS water extract
The 12 raw herbal medicines composing the GMSYS formula were purchased from a traditional herb market, Kwangmyungdang Medicinal Herbs (Ulsan, Republic of Korea). The 12 herbal medicines were authenticated by an expert taxonomist, Prof. Je-Hyun Lee, Dongguk University, Gyeongju, Republic of Korea. Voucher specimens were deposited at the K-herb Research Center, Korea Institute of Oriental Medicine (2012–KE45–1 ~ KE45–11). To obtain a water decoction of GMSYS, the 12 herbal medicines were mixed as shown in Table 1 (total weight = 5.0 kg, about 141.8 times the composition of a single dose) and extracted in distilled water at 100 °C for 2 h under pressure (98 kPa) using an electric extractor (COSMOS-660; Kyungseo Machine Co., Incheon, Korea). The extract was filtered using a standard sieve (No. 270, 53 μm; Chung Gye Sang Gong Sa, Seoul, Korea) and lyophilized to give a powder sample. The yield of GMSYS extract was about 19.4 % (970.4 g).
High-performance liquid chromatography (HPLC) analysis of GMSYS
Quantitative analysis of the GMSYS sample was performed using an LC-20A Prominence HPLC system (Shimadzu Corp., Kyoto, Japan) equipped with a solvent delivery unit, an on-line degasser, a column oven, an autosampler, and a photo diode array (PDA) detector. Data were acquired and processed using LabSolution software (version 5.54, SP3; Shimadzu Corp.). Separation was achieved on a SunFire C18 analytical column (250 × 4.6 mm; particle size 5 μm, Waters, Milford, MA, USA) as the stationary phase at a column temperature set to 40 °C. The mobile phases consisted of 0.1 % (v/v) formic acid in water (A) and 0.1 % (v/v) formic acid in acetonitrile (B). The gradient sequence and elution conditions were as follows: 5–60 % B for 0–30 min, 60–100 % B for 30–40 min, 100 % B for 40–45 min, 100–10 % B for 45–50 min, with a reequilibrium time of 10 min. The flow-rate was 1.0 mL/min, and the sample injection volume was 10 μL. For HPLC analysis, 200 mg of lyophilized GMSYS extract was dissolved in 20 mL of distilled water and then the solution diluted 10-fold for quantitative analysis of geniposide and paeoniflorin. Samples were filtered through a SmartPor GHP 0.2 μm syringe filter (Woongki Science, Seoul, Korea) before application onto the HPLC column.
Cell culture
The murine macrophage cell line, RAW 264.7, was obtained from the American Type Culture Collection (Rockville, MD). The cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco Inc., Grand Island, NY) supplemented with 5.5 % heat-inactivated fetal bovine serum (Gibco Inc.), penicillin (100 U/mL), and streptomycin (100 μg/mL) in a 5 % CO2 incubator at 37 °C.
Cytotoxicity assay
A cell viability assay was performed to determine the cytotoxicity of GMSYS using a Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan). RAW 264.7 macrophages were plated into a 96-well microplate at 3 × 103 cells/well and treated with various concentrations of GMSYS for 24 h. After incubation with CCK-8 reagent for 4 h, optical density (OD) at 450 nm was measured by using a Benchmark plus microplate reader (Bio-Rad Laboratories, Hercules, CA). The cell viability was calculated using the following equation:
NO assay
RAW 264.7 macrophages were pretreated with various concentrations of GMSYS (0, 250, 500, or 1000 μg/mL) for 4 h and stimulated with LPS (1 μg/mL) for an additional 20 h. NO synthesis was determined by measuring the accumulation of nitrite in the culture supernatant using a Griess Reagent System (Promega, Madison, WI). Briefly, an equal volume of supernatant and sulfanilamide solution was mixed and incubated for 10 min at room temperature, and then naphthylethylenediamine dihydrochloride solution was added. The mixture was incubated for an additional 5 min, and its absorbance was measured at 540 nm using a Benchmark plus microplate reader (Bio-Rad Laboratories). The nitrite concentration in the supernatants was determined from a standard curve generated with sodium nitrite.
ELISAs for PGE2, TNF-α, and IL-6
Production of PGE2, TNF-α, and IL-6 in the LPS-stimulated RAW 264.7 macrophage culture supernatants was measured using commercial ELISA kits from Cayman Chemical Co. (Ann Arbor, MI), Invitrogen (Carlsbad, CA), and BD Biosciences (Mountain View, CA), respectively.
RNA extraction and quantitative real-time polymerase chain reaction (RT-qPCR)
Total RNA was extracted using Trizol reagent (Invitrogen Life Sciences, Carlsbad, CA) according to the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized from 1 μg of total RNA using an iScript cDNA synthesis kit (Bio-Rad Laboratories). RT-qPCR was performed by using a Rotor Gene Q system (Qiagen, Hilden, Germany) and a reaction mixture that consisted of SYBR Green 2 × PCR Master Mix, cDNA template, and forward and reverse primers. The PCR protocol consisted of 35 cycles of denaturation at 95 °C for 15 sec, followed by 55 °C for 30 s to allow for extension and amplification of the target sequence. The relative levels of iNOS and COX-2 mRNA expression were normalized to that of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) using the 2-ΔΔCT method. The primer sequences used in this study are shown in Table 2.
Western blotting
RAW 264.7 macrophages were pretreated with GMSYS (0, 250, 500, or 1000 μg/mL) for 4 h and then treated with or without LPS (1 μg/mL) for 15 min (to detect phosphorylation of protein), or for 20 h (to detect total protein). Whole cell extract (WCE) was prepared by suspending the macrophages in an extraction lysis buffer (Sigma-Aldrich, St. Louis, MO) containing protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). Nuclear extract (NE) was isolated using NE-PER Nuclear and Cytoplasmic Extraction reagents (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer’s protocol. The protein concentration in the cell extracts was determined using a Bio-Rad Protein Assay Reagent (Bio-Rad Laboratories). Equal amounts of protein (30 μg) were resolved by 4–20 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane. The membrane was incubated with a solution of 5 % skim milk in Tris-buffered saline containing Tween 20 (TBST) to block nonspecific binding sites, followed by overnight incubation at 4 °C with an appropriate primary antibodies; anti-phospho-p38 MAPK, anti-phospho-ERK, anti-phospho-JNK (Cell Signaling, Danvers, MA), anti-NF-κB p65, anti-β-actin, anti-α-tubulin and anti-Lamin B (Santa Cruz Biotechnology, Dallas, TX). The membranes were washed three times with TBST, and then incubated with a corresponding horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA) for 1 h at room temperature. The membranes were washed three times with TBST again and then immunereactivity visualized using an enhanced chemiluminescence (ECL) (Thermo Fisher Scientific). Images were captured using Chemi-Doc (Bio-Rad Laboratories).
Statistical analyses
The data are expressed as the mean ± SEM. Data were analyzed using a one-way analysis of variance and Dunnett’s multiple comparisons test. P < 0.05 was considered to be significant.
Results
Identification and quantification of the marker components of GMSYS
We selected 11 components—including gallic acid, neomangiferin, chlorogenic acid, mangiferin, geniposide, paeoniflorin, berberine, liquiritin, nodakenin, glycyrrhizin, and atractylenolide III—as marker compounds of GMSYS. Calibration curves for the 11 marker components showed good linearity with correlation coefficients (r 2) ≥ 0.9996 in their various concentration ranges. Using optimized chromatography conditions, a three-dimensional chromatogram was obtained using HPLC–PDA detector, and the 11 compounds were eluted within 35 min (Fig. 1). The concentrations of gallic acid, neomangiferin, chlorogenic acid, mangiferin, geniposide, paeoniflorin, berberine, liquiritin, nodakenin, glycyrrhizin, and atractylenolide III were 0.58 ± 0.01, 0.45 ± 0.01, 0.90 ± 0.01, 1.99 ± 0.01, 8.37 ± 0.05, 8.91 ± 0.09, 2.01 ± 0.001, 0.94 ± 0.002, 1.43 ± 0.01, 0.81 ± 0.01, and 0.04 ± 0.001 mg/g, respectively.
Three-dimensional chromatogram of GMSYS by HPLC-PDA. The retention times of 11 marker compounds—gallic acid, neomangiferin, chlorogenic acid, mangiferin, geniposide, paeoniflorin, berberine, liquiritin, nodakenin, glycyrrhizin, and atractylenolide III—were approximately 6.32, 10.82, 12.63, 13.04, 13.53, 15.17, 15.35, 16.61, 17.27, 29.86, and 34.00 min, respectively
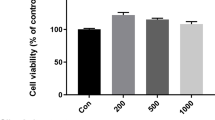
Cytotoxicity of GMSYS in RAW 264.7 macrophages
To determine the cytotoxicity of GMSYS, RAW 264.7 macrophages were treated with GMSYS at various concentrations ranging from 62.25 to 1000 μg/mL for 24 h, and a viability assay was conducted. As shown in Fig. 2, GMSYS did not produce any significant cytotoxicity up to 1000 μg/mL. Subsequent experiments were performed at nontoxic concentrations.
Effect of GMSYS on NO production and iNOS expression in LPS-stimulated RAW 264.7 macrophages
The level of NO in culture supernatant was measured using a Griess reaction. As shown in Fig. 3a, LPS treatment significantly increased the level of NO compared with vehicle-treated macrophages (P < 0.01). By contrast, LPS-induced NO production was markedly reduced in macrophages pretreated with GMSYS (500 or 1000 μg/mL). A positive control using N-methyl-l-arginine (l-NMMA, 100 μM) showed a significant decrease in LPS-induced NO production.
Effect of GMSYS on the production of NO and expression of iNOS in LPS-stimulated RAW 264.7 macrophages. The macrophages were pretreated with GMSYS for 4 h and then treated with LPS (1 μg/mL) for 20 h. a Collected supernatants were reacted with Griess reagent, and absorbance was measured at 540 nm. b Total RNA was isolated from the cell pellets and subjected to RT-qPCR to detect the expression of iNOS mRNA. Levels of iNOS mRNA expression were normalized to the expression of GAPDH mRNA. Bar graphs represent the means from three independent experiments. ## P < 0.01 vs vehicle-treated cells and * P < 0.05 or ** P < 0.01 vs LPS-treated cells
To investigate the cause of reduced NO production by GMSYS, the expression of iNOS by the macrophages was determined by RT-qPCR. LPS treatment of the macrophages significantly increased the expression of iNOS compared with vehicle-treated macrophages (P < 0.01). By contrast, GMSYS (500 or 1000 μg/mL) pretreatment significantly decreased the expression of LPS-induced iNOS (P < 0.01, Fig. 3b).
Effect of GMSYS on PGE2 production and COX-2 expression in LPS-stimulated RAW 264.7 macrophages
As shown in Fig. 4a, LPS-treated macrophages produced significantly increased levels of PGE2 compared with the vehicle-treated macrophages. By contrast, pretreatment with GMSYS (250, 500, or 1000 μg/mL) significantly decreased the level of PGE2 produced by the LPS-stimulated macrophages compared with macrophages treated with LPS alone (P < 0.01) (Fig. 3b). Indomethacin (2.5 ng/mL) was used as a positive control and markedly decreased PGE2 production by the stimulated macrophages.
Effect of GMSYS on the production of PGE2 and expression of COX-2 in LPS-stimulated RAW 264.7 macrophages. The macrophages were pretreated with GMSYS for 4 h and then treated with LPS (1 μg/mL) for 20 h. a PGE2 levels in supernatants were measures by ELISA. b Total RNA was isolated from the cell pellets and subjected to RT-qPCR to detect the expression of COX-2 mRNA. Levels of COX-2 mRNA expression were normalized to the expression of GAPDH mRNA. Bar graphs represent the means from three independent experiments. ## P < 0.01 vs vehicle-treated cells and ** P < 0.01 vs LPS-treated cells
To investigate whether the inhibitory effect of GMSYS on PGE2 production was related to the expression of COX-2, we measured the COX-2 mRNA expression using RT-qPCR. As shown in Fig. 4b, the expression of COX-2 was significantly induced by LPS stimulation (P < 0.01). By contrast, pretreatment of macrophages with GMSYS (250, 500, or 1000 μg/mL) markedly inhibited their LPS-induced COX-2 expression (P < 0.05, Fig. 4b).
Effect of GMSYS on the production of inflammatory cytokines in LPS-stimulated RAW 264.7 macrophages
To determine the effect of GMSYS on the production of proinflammatory cytokines TNF-α and IL-6, RAW 264.7 macrophages were pretreated with various concentrations of GMSYS (0, 250, 500, or 1000 μg/mL) for 4 h and then stimulated with LPS (1 μg/mL) for 20 h. As shown in Fig. 5, the levels of TNF-α and IL-6 were significantly increased in LPS-stimulated RAW 264.7 macrophages compared with vehicle-treated macrophages (P < 0.01). By contrast, pretreatment of the macrophages with GMSYS (250, 500, or 1000 μg/mL) significantly decreased the levels of TNF-α in a dose-dependent manner (P < 0.01) (Fig. 5a). LPS-induced IL-6 levels were also reduced by pretreatment of the macrophages with GMSYS (500 or 1000 μg/mL) (P < 0.01, Fig. 5b).
Effect of GMSYS on the production of TNF-α and IL-6 in LPS-stimulated RAW 264.7 macrophages. The macrophages were pretreated with GMSYS (0, 250, 500, or 1000 μg/mL) for 4 h and then treated with LPS (1 μg/mL) for 20 h. ELISAs were used to determined levels of (a) TNF-α and (b) IL-6 in collected supernatants. Bar graphs represent the means from three independent experiments. ## P < 0.01 vs vehicle-treated cells and ** P < 0.01 vs LPS-treated cells
Effect of GMSYS on MAPK phosphorylation in LPS-stimulated RAW 264.7 macrophages
To clarify the molecular mechanism of the anti-inflammatory effects of GMSYS, we analyzed the phosphorylation of p38, ERK, and JNK by Western blotting. As shown in Fig. 6a, GMSYS (250, 500, or 1000 μg/mL) decreased the expression of phosphorylated p38 in LPS-stimulated RAW 264.7 macrophages. In addition, the phosphorylation of ERK and JNK was also reduced by pretreatment of GMSYS (500 or 1000 μg/mL).
Effect of GMSYS on the phosphorylation of MAPKs and activation of NF-κB in LPS-stimulated RAW 264.7 macrophages. a The macrophages were pretreated with GMSYS (0, 250, 500, or 1000 μg/mL) for 2 h and then treated with LPS (1 μg/mL) for 15 min. Immunoblotting was used to detect the phosphorylation of p38 MAPK, ERK, and JNK in lysates prepared from the macrophages. b The macrophages were pretreated with GMSYS (0, 250, 500, or 1000 μg/mL) for 2 h and then treated with LPS (1 μg/mL) for 1 h. Immunoblotting of cytoplasmic and nuclear extracts prepared from the macrophages was used to detect the translocation of and subjected to immunoblotting for NF-κB p65. α-Tubulin and Lamin B were used as an internal control for cytoplasm and nucleus, respectively
Effect of GMSYS on NF-κB activation in LPS-stimulated RAW 264.7 macrophages
To investigate the mechanism by which GMSYS reduces LPS-induced inflammatory responses, the nuclear translocation of NF-κB p65 was assessed by Western blotting. As shown in Fig. 6b, nuclear NF-κB p65 was markedly increased in the nucleus of LPS-treated macrophages compared with the vehicle alone. By contrast, pretreatment with GMSYS reduced the nuclear translocation of NF-κB p65 in LPS-treated macrophages compared with those without such pretreatment.
Discussion
GMSYS, a traditional herbal formula comprising 12 different herbal medicines—Paeoniae Radix, Atractylodis Rhizoma Alba, Anemarrhenae Rhizoma, Lycii Radicis Cortex, Angelicae Gigantis Radix, Poria Sclerotium, Liriope Tuber, Rehmanniae Radix Crudus, Gardeniae Fructus, Phellodendri Cortex, Platycodi Radix, and Glycyrrhizae Radix et Rhizoma—has been used in Korea to treat dysmenorrhea, insomnia, and anxiety [12]. Eleven of these 12 herbal medicines exhibit anti-inflammatory properties; namely, Paeoniae Radix [21], Atractylodis Rhizoma Alba [22], Anemarrhenae Rhizoma [23], Lycii Radicis Cortex [24], Angelicae Gigantis Radix [25], Poria Sclerotium [26], Rehmanniae Radix Crudus [27], Gardeniae Fructus [28], Phellodendri Cortex [29], Platycodi Radix [30], and Glycyrrhizae Radix et Rhizoma [31].
In the present study, we analyzed 11 marker compounds found in an GMSYS—including gallic acid, neomangiferin, chlorogenic acid, mangiferin, geniposide, paeoniflorin, berberine, liquiritin, nodakenin, glycyrrhizin, and atractylenolide III—using HPLC-PDA detector. Nine of these compounds have anti-inflammatory effects; namely, gallic acid [32], chlorogenic acid [33], mangiferin [34], geniposide [35], paeoniflorin [36], berberine [37], nodakenin [38], glycyrrhizin [39], and atractylenolide III [40]. For these reasons, we predict that GMSYS, which contains these anti-inflammatory herbal medicines and components, has a preventive and therapeutic effect on inflammatory diseases. However, the anti-inflammatory effect and mechanisms of its action has not been thoroughly elucidated. Therefore, we sought to investigate the anti-inflammatory effect and molecular mechanisms of an extract of GMSYS in LPS-stimulated RAW 264.7 macrophages, to our knowledge for the first time. We found that GMSYS inhibited the release of inflammatory mediators such as NO and PGE2, and downregulated iNOS and COX-2 expression in LPS-stimulated macrophages. These findings suggest that GMSYS decreases NO and PGE2 levels by suppressing iNOS and COX-2 expression, respectively.
TNF-α and IL-6 have been implicated in autoimmune diseases including rheumatoid arthritis [41]. Therefore, the measurement of TNF-α and IL-6 production may suggest an anti-inflammatory effect. In the present study, we showed that GMSYS suppressed the production of TNF-α and IL-6 in LPS-stimulated RAW 264.7 macrophages.
MAPK represents a group of signaling molecules that appear to play critical roles in inflammatory processes. In particular, they control cellular responses to cytokines and play an important role in the modulation of NF-κB activity. LPS activates three molecules involved in MAPK cascades, p38, ERK, and JNK [42, 43]. To identify the signaling pathways involved in the GMSYS-mediated anti-inflammatory responses, we investigated the phosphorylation of these molecules by Western blotting. GMSYS diminished the phosphorylation of p38, ERK, and JNK in LPS-stimulated RAW 264.7 macrophages.
NF-κB plays an important role in various pathological states as a transcription factor for iNOS, TNF-α, and IL-1β, and is translocated into the nucleus by LPS stimulation [44]. In the present study, we found that GMSYS attenuated the nuclear translocation of p65 in LPS-stimulated RAW 264.7 macrophages. These findings suggest that GMSYS has an anti-inflammatory effect by inhibiting NF-κB activation.
Although synthetic drugs have been used for treatment of inflammatory diseases such as atherosclerosis, many of them exhibit varying degrees of adverse effects [45]. By contrast, an examination of herbal medicines may contribute to the discovery of novel drugs as potential anti-inflammatory agents with fewer side effects. Some traditional herbal formulas produce their anti-inflammatory effects by inhibiting the production of inflammatory mediators by blocking NF-kB activation in RAW 264.7 macrophages [46, 47]. These reports and our findings suggest that natural products including GMSYS could be used as anti-inflammatory agents with fewer adverse effects than synthetic drugs.
Conclusions
In conclusion, the present study showed that GMSYS inhibits the production and expression of NO, iNOS, PGE2, COX-2, TNF-α, and IL-6 in LPS-stimulated RAW 264.7 macrophages. These effects of GMSYS are related to the suppression of MAPK and NF-κB activation. Our results suggest that GMSYS should be considered as a source of potent anti-inflammatory candidates for the treatment or prevention of inflammatory diseases.
Abbreviations
CCK-8, cell counting kit-8; cDNA, complementary DNA; COX, cyclooxygenase; ECL, enhanced chemiluminescence; ERK, extracellular signal-regulated kinase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GMSYS, Gamisoyo-san; IL-6, interleukin-6; iNOS, inducible nitric oxide synthase; JNK, c-Jun N-terminal kinase; l-NMMA, N-methyl-l-arginine; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; NE, nuclear extract; NF-κB, nuclear transcription factor kappa B; NO, nitric oxide; NOS, nitric oxide synthase; OD, optical density; PDA, photo diode array; PGE2, prostaglandin E2; PVDF, polyvinylidene fluoride; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; TBST, tris-buffered saline containing Tween 20; TNF-α, tumor necrosis factor-α; WCE, whole cell extract
References
Reddy DB, Reddanna P. Chebulagic acid (CA) attenuates LPS-induced inflammation by suppressing NF-kappaB and MAPK activation in RAW 264.7 macrophages. Biochem Biophys Res Commun. 2009;381:112–7.
Fujiwara N, Kobayashi K. Macrophages in inflammation. Curr Drug Targets Inflamm Allergy. 2005;4:281–6.
Folino A, Losano G, Rastaldo R. Balance of nitric oxide and reactive oxygen species in myocardial reperfusion injury and protection. J Cardiovasc Pharmacol. 2013;62:567–75.
Nathan C. Nitric oxide as a secretory product of mammalian cells. FASEB J. 1992;6:3051–64.
Mitchell JA, Larkin S, Williams TJ. Cyclooxygenase-2: regulation and relevance in inflammation. Biochem Pharmacol. 1995;50:1535–42.
Laroux FS. Mechanisms of inflammation: the good, the bad and the ugly. Front Biosci. 2004;9:3156–62.
Blackwell TS, Christman JW. The role of nuclear factor-kappa B in cytokine gene regulation. Am J Respir Cell Mol Biol. 1997;17:3–9.
Ajizian SJ, English BK, Meals EA. Specific inhibitors of p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways block inducible nitric oxide synthase and tumor necrosis factor accumulation in murine macrophages stimulated with lipopolysaccharide and interferon-gamma. J Infect Dis. 1999;179:939–44.
Uto T, Fujii M, Hou DX. 6-(Methylsulfinyl)hexyl isothiocyanate suppresses inducible nitric oxide synthase expression through the inhibition of Janus kinase 2-mediated JNK pathway in lipopolysaccharide-activated murine macrophages. Biochem Pharmacol. 2005;70:1211–21.
Karin M, Delhase M. The I kappa B kinase (IKK) and NF-kappa B: key elements of proinflammatory signaling. Semin Immunol. 2000;12:85–98.
Baeuerle PA, Baltimore D. NF-kappa B: ten years after. Cell. 1996;87:13–20.
Kim JY, Kwak DH, Ju EJ, Kim SM, Lee DH, Keum KS, Lee SU, Jung KY, Seo BB, Choo YK. Effects of Gamisoyosan on in vitro fertilization and ovulation of stressed mice by electric shock. Arch Pharm Res. 2004;27:1168–76.
Lee JG, Shin BS, Lee YC, Park SW, Kim YH. Clinical effectiveness of the Kampo medicine kamishoyosan for adjunctive treatment of tardive dyskinesia in patients with schizophrenia: A 16-week open trial. Psychiatry Clin Neurosci. 2007;61:509–14.
Yamada K, Kanba S. Effectiveness of kamishoyosan for premenstrual dysphoric disorder: open-labeled pilot study. Psychiatry Clin Neurosci. 2007;61:323–5.
Terauchi M, Hiramitsu S, Akiyoshi M, Owa Y, Kato K, Obayashi S, Matsushima E, Kubota T. Effects of three Kampo formulae: Tokishakuyakusan (TJ-23), Kamishoyosan (TJ-24), and Keishibukuryogan (TJ-25) on Japanese peri- and postmenopausal women with sleep disturbances. Arch Gynecol Obstet. 2011;284:913–21.
Choi ES, Lee IS. Experimental study on effects of Soyosan and Kamisoyosan. J Orient Obstet Gynecol. 1996;9:41–53.
Hwang GS. Effects of Gamisoyosan (GS) on LDL oxidation in RAW 264.7 cell. Korean J Orient Prev Med Soc. 2001;5:134–43.
Lee SH, Lee JM, Cho JH, Lee CH, Jang JB, Lee KS. Antioxidant and Neuroprotective Effects of Gamisoyo-san. J Orient Obstet Gynecol. 2010;23:1–13.
Ko ES, Kang BC, Sung KH, Song IH, Kim UC, Kwon DI, Jung SM, Park JH. The clinical review on three cases of UL-syndrome induced by chronic stress. J Korean Orient Int Med. 2004;25:615–24.
Je YM, Yoo DY. Two case report of UL-syndrome treated with Gamisoyosan. Collection of Daejeon University (Thesis). 2011;19:187–93.
Lee B, Shin YW, Bae EA, Han SJ, Kim JS, Kang SS, Kim DH. Antiallergic effect of the root of Paeonia lactiflora and its constituents paeoniflorin and paeonol. Arch Pharm Res. 2008;31:445–50.
Yao CM, Yang XW. Bioactivity-guided isolation of polyacetylenes with inhibitory activity against NO production in LPS-activated RAW264.7 macrophages from the rhizomes of Atractylodes macrocephala. J Ethnopharmacol. 2014;151:791–9.
Lee EJ, Chung HJ, Pyee Y, Hong JY, Youn UJ, Seo EK, Lee SK. Suppression of inducible nitric oxide synthase expression by nyasol and broussonin A, two phenolic compounds from Anemarrhena asphodeloides, through NF-kB transcriptional regulation in vitro and in vivo. Chem Biodivers. 2014;11:749–59.
Lin CC, Chuang SC, Lin JM, Yang JJ. Evaluation of the antiinflammatory hepatoprotective and antioxidant activities of Lycium chinense from Taiwan. Phytomedicine. 1997;4:213–20.
Shin S, Jeon JH, Park D, Jang JY, Joo SS, Hwang BY, Choe SY, Kim YB. Anti-inflammatory effects of an ethanol extract of Angelica gigas in a Carrageenan-air pouch inflammation model. Exp Anim. 2009;58:431–6.
Jeong JW, Lee HH, Han MH, Kim GY, Hong SH, Park C, Choi YH. Ethanol extract of Poria cocos reduces the production of inflammatory mediators by suppressing the NF-kappaB signaling pathway in lipopolysaccharide-stimulated RAW 264.7 macrophages. BMC Complement Altern Med. 2014;14:101.
Liu CL, Cheng L, Ko CH, Wong CW, Cheng WH, Cheung DW, Leung PC, Fung KP, Bik-San Lau C. Bioassay-guided isolation of anti-inflammatory components from the root of Rehmannia glutinosa and its underlying mechanism via inhibition of iNOS pathway. J Ethnopharmacol. 2012;143:867–75.
Hong YJ, Yang KS. Anti-inflammatory activities of crocetin derivatives from processed Gardenia jasminoides. Arch Pharm Res. 2013;36:933–40.
Choi YY, Kim MH, Han JM, Hong J, Lee TH, Kim SH, Yang WM. The anti-inflammatory potential of Cortex Phellodendron in vivo and in vitro: down-regulation of NO and iNOS through suppression of NF-kB and MAPK activation. Int Immunopharmacol. 2014;19:214–20.
Jang MH, Kim CJ, Kim EH, Kim MG, Leem KH, Kim J. Effects of Platycodon grandiflorum on lipopolysaccharide-stimulated production of prostaglandin E2, nitric oxide, and interleukin-8 in mouse microglial BV2 cells. J Med Food. 2006;9:169–74.
Yang XL, Liu D, Bian K, Zhang DD. Study on in vitro anti-inflammatory activity of total flavonoids from Glycyrrhizae Radix et Rhizoma and its ingredients. Zhongguo Zhong Yao Za Zhi. 2013;38:99–104.
Chiu CS, Deng JS, Chang HY, Chen YC, Lee MM, Hou WC, Lee CY, Huang SS, Huang GJ. Antioxidant and anti-inflammatory properties of Taiwanese yam (Dioscorea japonica Thunb. var. pseudojaponica (Hayata) Yamam.) and its reference compounds. Food Chem. 2013;141:1087–96.
Hwang SJ, Kim YW, Park Y, Lee HJ, Kim KW. Anti-inflammatory effects of chlorogenic acid in lipopolysaccharide-stimulated RAW 264.7 cells. Inflamm Res. 2014;63:81–90.
Garrido G, Delgado R, Lemus Y, Rodríguez J, García D, Núñez-Sellés AJ. Protection against septic shock and suppression of tumor necrosis factor alpha and nitric oxide production on macrophages and microglia by a standard aqueous extract of Mangifera indica L. (VIMANG). Role of mangiferin isolated from the extract. Pharmacol Res. 2004;50:165–72.
Shi Q, Cao J, Fang L, Zhao H, Liu Z, Ran J, Zheng X, Li X, Zhou Y, Ge D, Zhang H, Wang L, Ran Y, Fu J. Geniposide suppresses LPS-induced nitric oxide, PGE2 and inflammatory cytokine by downregulating NF-kB, MAPK and AP-1 signaling pathways in macrophages. Int Immunopharmacol. 2014;20:298–306.
Jiang WL, Chen XG, Zhu HB, Gao YB, Tian JW, Fu FH. Paeoniflorin inhibits systemic inflammation and improves survival in experimental sepsis. Basic Clin Pharmacol Toxicol. 2009;105:64–71.
Jeong HW, Hsu KC, Lee JW, Ham M, Huh JY, Shin HJ, Kim WS, Kim JB. Berberine suppresses proinflammatory responses through AMPK activation in macrophages. Am J Physiol Endocrinol Metab. 2009;296:E955–964.
Rim HK, Cho W, Sung SH, Lee KT. Nodakenin suppresses lipopolysaccharide-induced inflammatory responses in macrophage cells by inhibiting tumor necrosis factor receptor-associated factor 6 and nuclear factor-kB pathways and protects mice from lethal endotoxin shock. J Pharmacol Exp Ther. 2012;342:654–64.
Fu Y, Zhou E, Wei Z, Liang D, Wang W, Wang T, Guo M, Zhang N, Yang Z. Glycyrrhizin inhibits the inflammatory response in mouse mammary epithelial cells and a mouse mastitis model. FEBS J. 2014;281:2543–57.
Li CQ, He LC, Jin JQ. Atractylenolide I and atractylenolide III inhibit lipopolysaccharide-induced TNF-alpha and NO production in macrophages. Phytother Res. 2007;21:347–53.
Wang Z, Jiang W, Zhang Z, Qian M, Du B. Nitidine chloride inhibits LPS-induced inflammatory cytokines production via MAPK and NF-kappaB pathway in RAW 264.7 cells. J Ethnopharmacol. 2012;144:145–50.
Vanden Berghe W, Plaisance S, Boone E, De Bosscher K, Schmitz ML, Fiers W, Haegeman G. p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways are required for nuclear factor-kappaB p65 transactivation mediated by tumor necrosis factor. J Biol Chem. 1998;273:3285–90.
Surh YJ, Chun KS, Cha HH, Han SS, Keum YS, Park KK, Lee SS. Molecular mechanisms underlying chemopreventive activities of anti-inflammatory phytochemicals: down-regulation of COX-2 and iNOS through suppression of NF-kappa B activation. Mutat Res. 2001;480–481:243–68.
Lee JW, Bae CJ, Choi YJ, Kim SI, Kim NH, Lee HJ, Kim SS, Kwon YS, Chun W. 3,4,5-Trihydroxycinnamic acid inhibits LPS-induced iNOS expression by suppressing NF-kB activation in BV2 microglial cells. Korean J Physiol Pharmacol. 2012;16:107–12.
Charo IF, Taub R. Anti-inflammatory therapeutics for the treatment of atherosclerosis. Nat Rev Drug Discov. 2011;10:365–76.
Kim MJ, Im KR, Yoon KS. Anti-inflammatory effects of YeongyoSeungma-tang. J Ethnopharmacol. 2009;126:377–81.
Chen G, Li KK, Fung CH, Liu CL, Wong HL, Leung PC, Ko CH. Er-Miao-San, a traditional herbal formula containing Rhizoma Atractylodis and Cortex Phellodendri inhibits inflammatory mediators in LPS-stimulated RAW264.7 macrophages through inhibition of NF-kB pathway and MAPKs activation. J Ethnopharmacol. 2014;154:711–8.
Acknowledgement
This research was supported by a grant for “Construction of Scientific Evidences for Herbal Medicine Formulas (K16251)” from the Korea Institute of Oriental Medicine.
Availability of data and materials
The data and materials of this article are included within the article.
Authors’ contributions
SEJ, HKS, and SJJ participated in the design of study. SEJ, OSK, SRY, CSS, YK, and SJJ performed experiments. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Jin, S.E., Kim, O.S., Yoo, SR. et al. Anti-inflammatory effect and action mechanisms of traditional herbal formula Gamisoyo-san in RAW 264.7 macrophages. BMC Complement Altern Med 16, 219 (2016). https://doi.org/10.1186/s12906-016-1197-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-016-1197-7