Abstract
Background
Epstein-Barr Virus (EBV) is a human oncogenic virus that can lead to cancer in lymphoid and epithelial cells and is one of the hypothesized causes of oral cavity lesions including oral squamous cell carcinoma (OSCC), but the etiological association remains undetermined. The present investigation aimed to explore the EBV presence, viral load, and EBV-encoded small RNA (EBER) sequence variation in tissue samples of patients with OSCC and other oral cavity lesions including oral lichen planus (OLP), and oral irritation fibroma (OIF).
Methods
In total, 88 oral cavity samples (23 with OSCC, 29 with OLP, and 36 with OIF diagnosis) were examined by Real-Time PCR technique and some of them were sequenced.
Results
Viral EBER sequence was detected in 6 out of the 23 OSCC (31.4%), 6 out of the 29 OLP (20.7%), and 3 out of the 36 OIF cases (8.3%). The mean EBV copy number was higher in OSCC samples (1.2 × 10−2 ± 1.3 × 10−2 copies/cell) compared to OLP (2.2 × 10−3 ± 2.6 × 10−3 copies/cell) and OIF (2.4 × 10−4 ± 2.0 × 10−4 copies/cell) samples, although this difference was not statistically significant (P = 0.318). The EBER gene was amplified and sequenced in 5 OSCC, 3 OLP, and 2 OIF samples with high EBV viral load. One OSCC, two OLP, and two OIF isolates showed different nucleotide variations compared with EBV-WT and AG876 prototype sequences: C6834T, C6870T, C6981T, C7085T, C7085G, and C7094T.
Conclusion
In our study the presence of more than one genome copies per tumor cell indicates the possible role of EBV infection in oral cancers. However, more studies should be conducted to clarify the role of EBV in OSCC carcinogenesis.
Similar content being viewed by others
Background
Oral squamous cell carcinoma (OSCC) is considered the most prevalent subset (90%) of oral cancer [1]. This malignancy has a survival rate of up to 80% if diagnosed early, compared to a survival rate of 20–30% if detected in late tumor stages [2]. It is the sixth most common malignancy worldwide [3]. The incidence and distribution of OSCC vary in different geographical locations and depends on various factors such as tobacco smoking, alcohol drinking, nutrition, and genetic components [4,5,6]. To perceive the full spectrum of oral carcinogenesis, the study of oral premalignant lesions (e.g., oral lichen planus) that carry a risk of malignant transformation is necessary. Oral lichen planus (OLP) is a common chronic inflammatory disorder with unknown etiology in which occurs as the result of abnormal T-cell-secreted cytokines in basal epithelial cells and resulting in either epithelial thickening or atrophy [7, 8]. The specific role of human tumor viruses including the Human Papilloma Virus (HPV) and Epstein-Barr Virus (EBV) in the development of OSCC and OLP continues to be a debated topic [9, 10]. Researchers proposed the first evidence for an association between OSCC and infectious agents, especially viruses, about two decades ago [11,12,13].
Epstein—Barr virus is a widespread human gamma herpes virus with oncogenic potential. It has been implicated in several malignant and benign lesions in the oral cavity and/or head and neck region including Burkitt’s lymphoma (BL), nasopharyngeal carcinoma (NPC), and oral hairy leukoplakia (OHL). The ability of EBV to infect oral and salivary glands epithelium and viral shedding via saliva indicate a possible role of the virus on OSCC pathogenesis. Despite the well-established impact of the EBV on BL and NPC, the influence of EBV in the etiology of OSCC remains uncertain. There have been several reports of EBV genome detection in both cancerous and non-cancerous oral epithelial tissues [14,15,16,17,18,19]. However, the association between EBV and OSCC remains controversial.
Regarding the role of carcinogenic viruses (e.g., EBV) in the development of cancer, several reports indicate that high viral load being proportional to the risk of viral oncogenesis and the number of viral genome copies per cell is an important indicator [20, 21]. For example, the higher EBV genome copy number increases the risk of Burkitt's lymphoma [22]. In addition, several lines of evidence indicate that certain variants of EBV with distinct polymorphisms in the EBV-encoded small RNA (EBER) region are strongly linked with NPC [23, 24]. Characterization of viral mutations in the EBER region seems important for EBV positive OSCC samples.
Thus, the facts reviewed above encouraged us to explore the presence of EBV genome in tissue samples of 88 patients with oral cavity lesions including OSCC, OLP, and oral irritation fibroma (OIF) and evaluate the positive samples in terms of viral copy number per cell. Moreover, genomic sequences of EBV in the EBER region were compared in 10 EBV positive samples isolated from OSCC, OLP, and OIF groups.
Methods
Patients and tissue samples
In this retrospective cross-sectional study, a total of 88 Formalin-fixed paraffin-embedded (FFPE) resection specimens were collected from the archives of the Pathology Department, School of Dentistry, affiliated to Babol University of Medical Sciences between 2015 -2017. All of the samples were subjected to histopathological diagnosis by an experienced pathologist. Out of the 88 FFPE samples, 23 had a histopathologic OSCC diagnosis, 29 and 36 samples had an OLP and an OIF diagnosis, respectively. The demographic and clinical characteristics of patients, including age, gender, anatomical localization of the specimens, and tumor grade were collected from subject's medical records. This study was approved by the Ethical Committee of Babol University of Medical Sciences (IR.MUBABOL.HRI.REC.1398.070), and for all subjects, written informed consent was obtained.
DNA extraction
Removal of paraffin from FFPE tissue sections was performed according to a previously described procedure [25]. Briefly, 5-μm-thick tissue slices were collected in a sterile nuclease-free microcentrifuge tube (10 sections from each tissue block and approximately 25 mg of FFPE sections). To prevent cross-contamination, a different microtome blade was utilized for each FFPE block. In order to dissolve the paraffin, the FFPE sections were incubated 3 times in 500 μL of xylene for 10 min at 60 °C and were subsequently washed with absolute ethanol. DNA was isolated using the DNA Extraction Mini Kit from Tissue (Yekta TajhizAzma, Tehran, Iran) according to the manufacturer’s instructions. In brief, for tissue digestion and protein removal, 200 μL of tissue lysis buffer and 20 μL of proteinase K (10 mg/mL) were added to each tube. Samples were subsequently incubated at 56 °C overnight. DNA cleanup was done by mini spin column (silica matrix) according to the manufacturer’s instructions. As an extraction negative control, sterile microcentrifuge tubes containing reaction mixtures were processed simultaneously with the tissue samples. The quality and quantity of extracted DNA were evaluated with a nanodrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) using the absorption ratio of 260/280 nm.
EBV quantitative real-time PCR
To detect and measure the amount of EBV viral load, quantitative real-time PCR was performed using a Rotor-Gene Q real-time PCR system (QIAGEN GmbH, Hilden, Germany), through the primer sets and a TaqMan probe specific for the EBV EBER gene according to a previously described procedure [26]. The EBV viral load was determined as the viral DNA copies/ half of the RNase P gene copy (each diploid cell contains two copies of RNase P gene), which normalizes viral copies to the number of cell equivalents as described previously [27]. Construction of plasmids containing cloned target sequences of EBV EBER and human RNase P gene (quantitative standards for real-time PCR) was done by gene synthesis service (Shanghai Gene ray Biotech Co., Ltd). Each reaction consisted of 100 ng of extracted DNA, 12.5 µL YTA qPCR Probe Master Mix (Yekta TajhizAzma, Tehran, Iran), 0.3 µM each primer, and 0.2 µM dual-labeled probe in a 25µL total reaction. In a biological hood, real-time PCR reaction mixes were prepared in a dedicated clean room. Each real-time PCR run included reaction mixtures without DNA template as a non-template control (NTC) and DNA extracted from supernatant of EBV-producing B-cell line (B95-8) as a positive control. To assess the sensitivity of the real-time PCR assay, a standard curve was generated using a tenfold dilution series of EBV EBER plasmid in genomic extracts obtained from EBV-negative FFPE samples.
Sequencing analysis of the EBV EBER gene
Ten EBV positive cases with high viral load isolated from OSCC, OLP, and OIF groups were subjected to sequence analysis. A 659 bp fragment of the EBER from nucleotide positions 6758–7416 (based on wild type EBV; GenBank accession number NC_007605) was amplified by PCR according to a previously described procedure [23]. The purified PCR products were sent to Bioneer Corporation (Daedeok-gu, Daejeon, South Korea) for dideoxy-DNA sequencing. Sequencing was performed on both strands using the same primers as in PCR. The raw sequencing data were aligned and evaluated using CLC Genomics Workbench (CLC Bio, Aarhus, Denmark). The representative EBV EBER nucleotide sequences reported in this study were deposited in the GenBank with accession numbers MN921212 to MN921221. The maximum-likelihood method was utilized for the construction of a phylogenetic tree of the EBV EBER region (nucleotide positions 6758–7416) for all of the sequences, using MEGA7 software. Statistical significance of the phylogenetic tree was checked by the bootstrap method (1,000 replicates). Besides for out-group, we used a viral sequence other than EBV (Herpes Simplex Virus Type 1).
Statistical analysis
SPSS version 23 was used to conduct the statistical analysis (SPSS Inc., Chicago, IL, USA). The chi-square analysis was used to analyze differences in proportions between categorical variables. The Kolmogorov–Smirnov test was used to determine if the variables had a normal distribution. We used the independent-samples T-test and the Kruskal–Wallis test for analysis of continuous variables. Statistical significance was described as a P value of less than 0.05.
Results
Study patient characteristics
All 88 enrolled subjects (35 men and 53 women, mean age, 51.6 ± 17 years; range, 12–87) were categorized in to three groups: (1) subjects with histopathological confirmed malignant OSCC (n = 23, 13 men and 10 women; mean age, 68.6 ± 13.4 years) (2) those with oral pre-malignant OLP disorder (n = 29, 11 men and 18 women; mean age, 46.7 ± 12 years) and (3) subjects with non-neoplastic OIF (n = 36, 11 men and 25 women; mean age, 44.1 ± 14.9). The demographic and clinical characteristics of the participants are displayed in Table 1. The patient's samples had been taken from different sites in the oral cavity including buccal mucosa (44.3%), gingiva & alveolar mucosa (14.8%), tongue (12.5%), labial mucosa (11.4%), palate (4.5%), and floor of mouth (1.1%). The anatomical localization of 10 samples was unknown.
Detection and quantitation of EBV
DNA quality and integrity of all included samples were verified by successful amplification of the human RNase P gene. Samples with cycle threshold (Ct) values ≥ 30 for human RNase P were considered to have inadequate DNA quality and therefore were re-extracted until RNase P amplification could be achieved with Ct values < 30. The results from EBV detection revealed the presence of the viral EBER gene in a total of 15 (17%) out of the 88 tested samples (4 men and 11 women; mean age, 55.7 ± 15.9 years). No significant differences were found between EBV positivity and gender (P = 0.386). Also, no statistically significant difference between patients’ mean age and EBV positivity was seen (P = 0.570). Of the 23 OSCCs, EBV DNA was detected in 6 tumors (26.1%). In detail, out of six OSCC positive samples, EBV DNA was detected in three (50%) gingiva & alveolar mucosa, one (16.6%) of buccal mucosa, one (16.6%) of the floor of the mouth. The anatomical localization of one EBV positive OSCC sample was unknown. Regarding the grade of tumor in the OSCC group, EBV genome was detected in 36.4% (4/11) of grade I, and 14.3% (1/7) of grade II tumors. The grade of the tumor was unknown in one EBV positive OSCC. Of the 29 OLPs, EBV DNA was detected in 6 samples (20.7%). In the OLP group, EBV DNA was detected in four (66.6%) of buccal mucosa, one (16.6%) of labial mucosa, and one (16.6%) of the palate. Epstein—Barr virus DNA was detected in oral specimens of only 3 out of the 36 OIF cases (8.3%), specifically, two (5.5%) gingiva & alveolar mucosa, and one (16.6%) of the buccal mucosa. There was no significant difference in EBV positivity between the three study groups (OSCC, OLP, and OIF) (P = 0.18) (Table 1).
The EBV DNA load was determined as the viral copy number per cell using a proven single-copy gene, human RNase P. In EBV positive samples, the mean EBV copy number was higher in OSCC samples (1.2 × 10−2 ± 1.3 × 10−2 copies/cell) compared to OLP (2.2 × 10−3 ± 2.6 × 10−3 copies/cell) and OIF (2.4 × 10−4 ± 2.0 × 10−4 copies/cell) samples, although this difference was not statistically significant (P = 0.318) (Fig. 1A). In terms of the various site of lesions in the oral cavity the mean EBV copy number in the buccal mucosa, gingiva & alveolar mucosa, and other sites were 2.3 × 10−3 ± 3.5 × 10−3, 9.3 × 10−3 ± 1.6 × 10−2, and 7.9 × 10−3 ± 1.1 × 10−2 copies/cell, respectively. There was no statistically significant difference between the various site of lesions and EBV DNA load (P = 0.596) (Fig. 1B). Regarding the grade of OSCC tumor the mean EBV copy number in grade I tumors was 1.7 × 10−2 ± 1.5 × 10−2, and a very low level of EBV DNA (7.0 × 10−5 copies/cell) was found in only 1 EBV positive grade II tumor.
EBER sequence variations
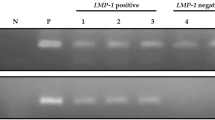
The EBER gene was amplified and sequenced in 5 OSCC, 3 OLP, and 2 OIF samples with high EBV viral load. Comparison of nucleotide sequences of the coding region of EBER2 (6956–7128) as well as non-coding region between EBER1 and EBER2 (6796–6955) with prototype EBV-WT reference sequence was performed. The sequence variations are shown in Table 2. Mutations at the seven nucleotides have been regarded as the type-specific mutations (6808, 6884, 6886, 6911, 6927, 6944, and 7123), and sequences identical to EBV-WT at the seven positions are classified as type 1 EBER sequences, whereas those identical to AG876 at the seven positions are classified as type 2 EBER sequences [28, 29]. In the present study, all the 10 isolates are classified as type 1 EBER sequence according to the similarity in the aforementioned seven nucleotide positions. One OSCC, two OLP, and two OIF isolates showed different nucleotide variations compared with EBV-WT and AG876 prototype sequences: C6834T, C6870T, C6981T, C7085T, C7085G, and C7094T. The 10 sequences of the EBER region (nucleotide positions 6758–7416) from all of the patients and the 3 EBER sequences of reported EBV genomes (EBV-WT, AG876, and GD1 strain) were used to make a phylogenetic tree (Fig. 2). The phylogenetic tree is labeled with patient group (OSCC, OLP, and OIF), and accession number. No clustering according to the patient group was observed among the sequences.
Phylogenetic tree for sequences of ten selected EBV positive patients in the EBER region (nucleotide positions 6758–7416) with MEGA (version 7.0) software. The patient group (OSCC, OLP and OIF), and accession number are listed. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model [55]. The tree with the highest log likelihood (-2588.74) is shown
Discussion
Oral cancer is the sixth most frequent cancer in the human population, with OSCC being the most common type [30]. Oral squamous cell carcinoma can be caused by a variety of risk factors, including oncogenic viruses [31]. In different parts of the world, various EBV prevalence rate on OSCC samples has been reported [14, 15, 32,33,34]. Different research groups have different perspectives on the role of EBV in OSCC. This inconsistency may be as a result of using different methods in studies, geographic and ethnic variations and also the effects of the other risk factors.
In several studies with no evidence of EBV detection in OSCC samples, the "hit and run" theory was proposed. According to this theory, viral DNA is only necessary for malignancy induction and then vanishes during the host's uncontrolled cell cycles [35]. In some studies, EBV genomic sequences has been isolated from OSCC samples using PCR technique [18, 36]. In addition, EBV EBER was found to be overexpressed in extreme epithelial dysplasia and linked to carcinogenesis [37]. In this way, we used the EBER gene to evaluate the presence of EBV in the OSCC samples. In the present study, the patient's samples were taken from different sites in the oral cavity. The results from EBV detection revealed the existence of the viral EBER gene in 17% of samples (31.4% of OSCC, 20.7% of OLP and 8.3% of OIF). According to an investigation, EBV infection in OSCCs was detected by PCR in 52.8 percent of cases and by in-situ hybridization (ISH) EBER was found in 27.5 percent of cases [38]. In another study, the researchers found positivity for EBV infection in all of 36 (100%) OSCCs, 7 out of 9 (77.8%) pre-cancerous lesions, and 1of 12 (8.3%) normal mucosa [39], which show the high frequency of EBV infection in OSCC samples. A Taiwanese research team using an EBV genomic microarray found a high rate of EBV infection (82.5%) in biopsy specimens from 57 OSCC cases [40]. Reporting the high prevalence of EBV infection in their research can be as a result of using fresh tissue samples and higher technical sensitivity. The other two studies which evaluated FFPE samples with PCR method, also declared that the EBV prevalence was higher in OSCC in comparison with healthy individuals [18, 41].
In contrast, Lamaroon and colleagues in a study conducted in Thailand didn’t consider EBV as an OSCC risk factor since the EBER gene was not detectable in their 24 cases of OSCC [42]. Similarly, Kerishnan et al. revealed that their results didn’t demonstrate any association between EBV infection and OSCC [43]. About the location of malignancies, we didn’t see any statistically significant difference between the various sites of lesions and EBV DNA detection. Our findings were consistent with two other studies [15, 44]. Furthermore, no statistically significant difference between patients’ mean age and EBV positivity was seen in our findings. Comparable to our results there was also no link between EBV viral load and patient age in positive cases in Saravani et al. investigation [45].
The presence of more than one genome copies per tumor cell, which is generally seen in BL and NPC, suggests direct EBV oncogenic role. In the current investigation, in accordance with Goldenberg et al. [46], less than one EBV genome per cell has been detected in OSCC samples. Low copy numbers of virus genome per tumor cell, suggests persistence of EBV as a passenger virus in the oral cavity without obvious pathological consequence. However, several studies have demonstrated substantially higher levels of EBV genome in OSCC compared non-cancerous tissues [47,48,49,50].
In this study, ten EBV positive cases with high viral load isolated from OSCC, OLP, and OIF groups were subjected to EBV mutation analysis of spacer and EBER2 sequences to gain a true picture of EBER variant polymorphisms. One OSCC, two OLP, and two OIF isolates showed different nucleotide variations compared with EBV-WT and AG876 prototype sequences. However, no consensus mutations were found among the isolates regarding three main EBER variants (EB-6 m, EB-8 m, and EB-10 m). Three significant EBER gene variations denoted as EB-6 m, EB-8 m, and EB-10 m, were found in a study on northern Chinese isolates from the NPC non-endemic area, which EB-8 m and EB-10 m, had six common mutations at the EBER2 coding region [51]. EBV-encoded small RNA gene polymorphism has received little research because it is widely assumed that the coding sequences of EBER1 and EBER2 are conserved between EBV strains [28, 29]. However, this viral gene has a small amount of sequence variation, which has been related to the EBV Type 1/Type 2 status [29, 52]. In the present study, all the 10 isolates are classified as EBV Type 1, based on the similarity to EBV-WT at the EBER sequence. Sequence analysis of EBV strains B95-8, P3HR-1, and AG876 revealed two patterns of EBER variants, B95-8 and AG876/P3HR-1, with the majority of type-specific mutations occurring in the 161-bp noncoding spacer region [29]. In another study by Xiaoshi et al., EBERs from NPC tissue possessed the basic characteristics of type 1 EBV, as well as 8 base-pair substitutions and 2 base pair deletions in EBER-2 [53].
Taken together, current study had some limitations including, small sample size, lack of fresh or non-paraffin samples, and no evaluation of non-cancerous tumor margin.
Conclusion
This study found that EBV is more common in oral lesions like OSCC and OLP than in OIF with a low copy number of viral genomes. The presence of EBV in our samples can show the indirect role of this virus in oral disease pathogenesis, that they use a "hit and run" process, or that they are only involved in a small percentage of cases. However, with the limitation of the findings, many issues about the molecular pathways underlying EBV infection-induced carcinogenesis remain unanswered, so prospective follow-up studies are needed to investigate the mechanisms by which EBV promotes carcinogenesis. Also, epidemiological and molecular investigations, especially on oral fresh biopsy samples and in case–control setting should be done to ascertain the role of EBV in oral carcinogenesis.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- EBV:
-
Epstein-Barr Virus
- OSCC:
-
Oral squamous cell carcinoma
- EBER:
-
EBV-encoded small RNA
- OLP:
-
Oral lichen planus
- OIF:
-
Oral irritation fibroma
- HPV:
-
Human Papilloma Virus
- BL:
-
Burkitt’s lymphoma
- NPC:
-
Nasopharyngeal carcinoma
- OHL:
-
Oral hairy leukoplakia
- FFPE:
-
Formalin-fixed paraffin-embedded
- NTC:
-
Non-template control
- CT:
-
Cycle threshold
- WT:
-
Wild type
References
Massano J, Regateiro FS, Januário G, Ferreira A. Oral squamous cell carcinoma: review of prognostic and predictive factors. Oral Surg Oral Med Oral Pathol Oral Radiol Endodontol. 2006;102(1):67–76.
Dumache R. Early diagnosis of oral squamous cell carcinoma by Salivary microRNAs. Clin Lab. 2017;63(11):1771–6.
Chiou S-H, Yu C-C, Huang C-Y, Lin S-C, Liu C-J, Tsai T-H, et al. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res. 2008;14(13):4085–95.
Reichart P, Nguyen X. Betel quid chewing, oral cancer and other oral mucosal diseases in Vietnam: a review. J Oral Pathol Med. 2008;37(9):511–4.
Marur S, Forastiere AA, editors. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clinic Proceedings; 2016: Elsevier.
Goldstein BY, Chang S-C, Hashibe M, La Vecchia C, Zhang Z-F. Alcohol consumption and cancer of the oral cavity and pharynx from 1988 to 2009: an update. ECP. 2010;19(6):431.
Khan A, Farah CS, Savage NW, Walsh LJ, Harbrow DJ, Sugerman PB. Th1 cytokines in oral lichen planus. J Oral Pathol Med. 2003;32(2):77–83.
Sugerman PB, Savage NW, Zhou X, Walsh LJ, Bigby M. Oral lichen planus. Clin Dermatol. 2000;18(5):533–9.
Sand L, Jalouli J. Viruses and oral cancer. Is there a link? Microbes and infection. 2014;16(5):371–8.
Speicher DJ, Ramirez-Amador V, Dittmer DP, Webster-Cyriaque J, Goodman MT, Moscicki AB. Viral infections associated with oral cancers and diseases in the context of HIV: a workshop report. Oral Dis. 2016;22:181–92.
Scully C. Oral squamous cell carcinoma; from an hypothesis about a virus, to concern about possible sexual transmission. Oral Oncol. 2002;38(3):227–34.
Campisi G, Panzarella V, Giuliani M, Lajolo C, Di Fede O, Falaschini S, et al. Human papillomavirus: Its identikit and controversial role in oral oncogenesis, premalignant and malignant lesions. Int J Oncol. 2007;30(4):813–23.
Andrews E, Seaman WT, Webster-Cyriaque J. Oropharyngeal carcinoma in non-smokers and non-drinkers: a role for HPV. Oral Oncol. 2009;45(6):486–91.
Shimakage M, Horii K, Tempaku A, Kakudo K, Shirasaka T, Sasagawa T. Association of Epstein-Barr virus with oral cancers. Hum Pathol. 2002;33(6):608–14.
Sand LP, Jalouli J, Larsson P-A, Hirsch J-M. Prevalence of Epstein-Barr virus in oral squamous cell carcinoma, oral lichen planus, and normal oral mucosa. Oral Surg Oral Med Oral Pathol Oral Radiol Endodontol. 2002;93(5):586–92.
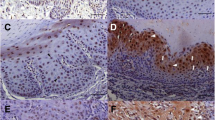
Shamaa AA, Zyada MM, Wagner M, Awad SS, Osman MM, Azeem AAA. The significance of Epstein Barr virus (EBV) & DNA topoisomerase II alpha (DNA-Topo II alpha) immunoreactivity in normal oral mucosa, oral epithelial dysplasia (OED) and oral squamous cell carcinoma (OSCC). Diagn Pathol. 2008;3(1):1–13.
Began J, Jiménez Y, Murillo J, Poveda R, Diaz JM, Sanchis J, et al. Epstein-Barr virus in oral proliferative verrucous leukoplakia and squamous cell carcinoma: A preliminary study. Med Oral Patol Oral Y Cirugia Bucal. 2008;13(2):110.
Kis A, Fehér E, Gáll T, Tar I, Boda R, Tóth ED, et al. Epstein-Barr virus prevalence in oral squamous cell cancer and in potentially malignant oral disorders in an eastern Hungarian population. Eur J Oral Sci. 2009;117(5):536–40.
Jiang R, Gu X, Moore-Medlin TN, Nathan C-A, Hutt-Fletcher LM. Oral dysplasia and squamous cell carcinoma: correlation between increased expression of CD21, Epstein-Barr virus and CK19. Oral Oncol. 2012;48(9):836–41.
Hsieh P-P, Tung C-L, Chan AB-W, Liao J-B, Wang J-S, Tseng H-H, et al. EBV viral load in tumor tissue is an important prognostic indicator for nasal NK/T-cell lymphoma. American journal of clinical pathology. 2007;128(4):579–84.
Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernández‐Figueras MT, et al. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. International journal of cancer. 2009;125(6):1243–9.
Rosemarie Q, Sugden B. Epstein-Barr Virus: how its lytic phase contributes to oncogenesis. Microorganisms. 2020;8(11):1824.
Hui KF, Chan TF, Yang W, Shen JJ, Lam KP, Kwok H, et al. High risk Epstein-Barr virus variants characterized by distinct polymorphisms in the EBER locus are strongly associated with nasopharyngeal carcinoma. Int J Cancer. 2019;144(12):3031–42.
Palser AL, Grayson NE, White RE, Corton C, Correia S, Ba abdullah MM, et al. Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J Virol. 2015;89(10):5222–37.
Yahyapour Y, Shamsi-Shahrabadi M, Mahmoudi M, Siadati S, Shahryar SS, Shokri-Shirvani J, et al. Evaluation of human papilloma virus infection in patients with esophageal squamous cell carcinoma from the Caspian Sea area, north of Iran. Asian Pac J Cancer Prev. 2012;13(4):1261–6.
Ling PD, Vilchez RA, Keitel WA, Poston DG, Peng RS, White ZS, et al. Epstein-Barr virus DNA loads in adult human immunodeficiency virus type 1-infected patients receiving highly active antiretroviral therapy. Clin Infect Dis. 2003;37(9):1244–9.
Sadeghi F, Salehi-Vaziri M, Ghodsi SM, Alizadeh A, Bokharaei-Salim F, Saroukalaei ST, et al. Prevalence of JC polyomavirus large T antigen sequences among Iranian patients with central nervous system tumors. Adv Virol. 2015;160(1):61–8.
Lin J-C, Lin S-C, De BK, Chan W-P, Evatt BL. Precision of genotyping of Epstein-Barr virus by polymerase chain reaction using three gene loci (EBNA-2, EBNA-3C, and EBER): predominance of type A virus associated with Hodgkin’s disease. Blood. 1993;81(12):3372–81.
Arrand JR, Young L, Tugwood J. Two families of sequences in the small RNA-encoding region of Epstein-Barr virus (EBV) correlate with EBV types A and B. J Virol. 1989;63(2):983–6.
Dhanuthai K, Rojanawatsirivej S, Thosaporn W, Kintarak S, Subarnbhesaj A, Darling M, et al. Oral cancer: A multicenter study. Medicina oral, patologia oral y cirugia bucal. 2018;23(1):e23.
Orbak R, Bayraktar C, Kavrut F, Gündogdu C. Poor oral hygiene and dental trauma as the precipitating factors of squamous cell carcinoma. Oral Oncol Extra. 2005;41(6):109–13.
Kobayashi I, Shima K, Saito I, Kiyoshima T, Matsuo K, Ozeki S, et al. Prevalence of Epstein-Barr virus in oral squamous cell carcinoma. J Pathol. 1999;189(1):34–9.
Gonzalez-Moles M, Gutierrez J, Rodriguez M, Ruiz-Avila I, Rodriguez-Archilla A. Epstein-Barr virus latent membrane protein-1 (LMP-1) expression in oral squamous cell carcinoma. Laryngoscope. 2002;112(3):482–7.
D’Costa J, Saranath D, Sanghvi V, Mehta AR. Epstein-Barr virus in tobacco-induced oral cancers and oral lesions in patients from India. J Oral Pathol Med. 1998;27(2):78–82.
Hermann R, Füzesi L, Pradier O, Christiansen H, Schmidberger H. Presence of human papillomavirus-18 and Epstein–Barr virus in a squamous cell carcinoma of the tongue in a 20-year-old patient. Case report and review of the current literature. Cancer/Radiothérapie. 2004;8(4):262–5.
Tsuhako K, Nakazato I, Miyagi J, Iwamasa T, Arasaki A, Hiratsuka H, et al. Comparative study of oral squamous cell carcinoma in Okinawa, Southern Japan and Sapporo in Hokkaido, Northern Japan; with special reference to human papillomavirus and Epstein-Barr virus infection. J Oral Pathol Med. 2000;29(2):70–9.
Shimizu N, Tanabe-Tochikura A, Kuroiwa Y, Takada K. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt’s lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J Virol. 1994;68(9):6069–73.
Horiuchi K, Mishima K, Ichijima K, Sugimura M, Ishida T, Kirita T. Epstein-Barr virus in the proliferative diseases of squamous epithelium in the oral cavity. Oral Surg Oral Med Oral Pathol Oral Radiol Endodontol. 1995;79(1):57–63.
Cruz I, Van den Brule A, Steenbergen R, Snijders P, Meijer C, Walboomers J, et al. Prevalence of Epstein–Barr virus in oral squamous cell carcinomas, premalignant lesions and normal mucosa—a study using the polymerase chain reaction. Oral Oncol. 1997;33(3):182–8.
Tsang NM, Chang KP, Lin SY, Hao SP, Tseng CK, Kuo Tt, et al. Detection of Epstein‐Barr Virus–Derived Latent Membrane Protein‐1 Gene in Various Head and Neck Cancers: Is It Specific for Nasopharyngeal Carcinoma? The Laryngoscope. 2003;113(6):1050–4.
Prathyusha MM, Kattappagari KK, Chowdary D, Shekar PC, Alivelu D, Reddy BVR. A study on association of epstein barr virus in oral squamous cell carcinoma using polymerase chain reaction technique. J Dr NTR Univ Health Sci. 2019;8(4):233.
Iamaroon A, Khemaleelakul U, Pongsiriwet S, Pintong J. Co-expression of p53 and Ki67 and lack of EBV expression in oral squamous cell carcinoma. J Oral Pathol Med. 2004;33(1):30–6.
Kerishnan JP, Mah MK, Mohd Fawzi NA, Ramanathan A, Lim GS, Abdul Aziz A, et al. The association between Epstein-Barr virus (EBV) Past Infection with the risk of oral squamous cell carcinoma (OSCC). Sains Malaysiana. 2019;48(9):1969–74.
Maeda T, Hiranuma H, Matsumura S, Furukawa S, Fuchihata H. Epstein-Barr virus infection and response to radiotherapy in squamous cell carcinoma of the oral cavity. Cancer Lett. 1998;125(1–2):25–30.
Saravani S, Miri-Moghaddam E, Sanadgol N, Kadeh H, Nazeri MR. Human herpesvirus-6 and Epstein-Barr virus infections at different histopathological grades of oral squamous cell carcinomas. Int J Prev Med. 2014;5(10):1231.
Goldenberg D, Benoit NE, Begum S, Westra WH, Cohen Y, Koch WM, et al. Epstein-Barr virus in head and neck cancer assessed by quantitative polymerase chain reaction. Laryngoscope. 2004;114(6):1027–31.
Bagan JV, Jiménez Y, Murillo J, Poveda R, Díaz JM, Gavaldá C, et al. Epstein-Barr virus in oral proliferative verrucous leukoplakia and squamous cell carcinoma: A preliminary study. Med Oral Patol Oral Cir Bucal. 2008;13(2):E110–3.
Cruz I, Van den Brule AJ, Steenbergen RD, Snijders PJ, Meijer CJ, Walboomers JM, et al. Prevalence of Epstein-Barr virus in oral squamous cell carcinomas, premalignant lesions and normal mucosa–a study using the polymerase chain reaction. Oral Oncol. 1997;33(3):182–8.
Jiang R, Ekshyyan O, Moore-Medlin T, Rong X, Nathan S, Gu X, et al. Association between human papilloma virus/Epstein-Barr virus coinfection and oral carcinogenesis. J Oral Pathol Med. 2015;44(1):28–36.
Sand LP, Jalouli J, Larsson PA, Hirsch JM. Prevalence of Epstein-Barr virus in oral squamous cell carcinoma, oral lichen planus, and normal oral mucosa. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2002;93(5):586–92.
Wang Y, Zhang X, Chao Y, Jia Y, Xing X, Luo B. New variations of Epstein-Barr virus-encoded small RNA genes in nasopharyngeal carcinomas, gastric carcinomas, and healthy donors in northern China. J Med Virol. 2010;82(5):829–36.
Schuster V, Ott G, Seidenspinner S, Kreth HW. Common Epstein-Barr virus (EBV) type-1 variant strains in both malignant and benign EBV-associated disorders. 1996.
Xiaoshi Z, Linjie Z, Ruhua Z, Haiqiang M, Jianjing C, Yixin Z. Relationship between polymorphism of Epstein-Barrvirus-encoded small RNAs and nasopharyngeal carcinomas. J Pract Oncol. 2001;16(5):301–3.
Dolan A, Addison C, Gatherer D, Davison AJ, McGeoch DJ. The genome of Epstein-Barr virus type 2 strain AG876. Virology. 2006;350(1):164–70.
Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–26.
Acknowledgements
We would like to express our appreciation to the directors and staff of Oral and Maxillofacial Pathology Department, School of Dental Medicine, Babol University of Medical Sciences for their collaboration in sample collection. This study was financially supported by a grant from Babol University of Medical Sciences (Project code: 9807346).
Funding
No funding.
Author information
Authors and Affiliations
Contributions
AZ: prepared the writing and drafting. YY: revised the writing and drafting. MM: Collaborate on collecting samples. MC: Statistical data analysis and draw statistical charts. FS: supervised the data and approved the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethical Committee of Babol University of Medical Sciences (IR.MUBABOL.HRI.REC.1398.070), and for all subjects, written informed consent was obtained.
Competing interests
The authors declare that they have no compete of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zebardast, A., Yahyapour, Y., Majidi, M.S. et al. Detection of Epstein-Barr virus encoded small RNA genes in oral squamous cell carcinoma and non-cancerous oral cavity samples. BMC Oral Health 21, 502 (2021). https://doi.org/10.1186/s12903-021-01867-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12903-021-01867-8