Abstract
Background
The VicRK two-component signalling system regulates virulence and cariogenicity in Streptococcus mutans (S. mutans). The purpose of this study was to explore the genetic polymorphisms of the vicR and vicK genes, which are associated with dental caries in children with S. mutans.
Methods
In this study, 121 (from each group) clinical S. mutans strains were isolated from caries-free children and children with high-severity caries to sequence the vicR and vicK genes. Genomic DNA was extracted from S. mutans strains and amplified using PCR. The PCR products were purified and sequenced. A chi-squared test and ABI Variant Reporter software were used to analyse the sequencing results.
Results
The 242 clinically isolated S. mutans strains contained the full-length vicR and vicK genes. No nucleotide sequence insertions or deletions were observed in the two genes. Four silent point mutations were identified in the vicR genes, and no missense mutations could be detected. Forty-one mutations were identified in the vicK genes. In addition to 32 silent mutations, 9 missense mutations at the 173, 337, 470, 1051, 1132, 1258, 1260, 1277, and 1348 bp positions were found. The distribution frequencies of the missense mutations were not significantly different between the groups, except for the C470T mutation. The frequency of the C470T missense mutation was higher in the high-severity caries group than in the caries-free group.
Conclusions
vicR sequences are highly conserved in S. mutans clinical isolates. The locus 470 missense mutation of the vicK gene may be related to caries in children with S. mutans.
Similar content being viewed by others
Background
The human oral microbial ecosystem is one of the most complex bacterial communities known. Approximately 700 bacterial species are estimated to exist in the oral cavity [1, 2]. Among these bacteria, Streptococcus mutans (S. mutans) is one of the few species that have been consistently linked with dental caries [3, 4]. From the initial colonization stages onward, S. mutans undergoes continuous dynamic challenges, such as changes in pH, temperature, and nutrient levels, to which it must respond and adapt. Generally, in bacteria, two-component regulatory systems (TCRSs) are used as “molecular switches” in response to environmental changes [5, 6]. These regulatory networks are essential for bacterial adaptation, survival, and virulence.
Based on the genome sequence, 13 TCRSs have been identified in S. mutans UA159 [7]. One of these TCRSs, the VicRK signal transduction system, affects various virulence attributes of S. mutans [8, 9]. The system is composed of a histidine kinase sensor protein (VicK) located in the membrane and a cytoplasmic response regulator protein (VicR). Through phosphorylation reactions, extracellular signals are sensed by VicK, and then the VicK histidine kinase transmits the message to VicR, which modulates gene expression [10, 11].
vic genes regulate the expression of several virulence-associated genes that affect synthesis and adhesion to polysaccharides, including gtfBCD, ftf, and gbpB [12]. Moreover, compared with the wild-type UA159 strain, strains without vicK form aberrant biofilms, with a reduced rate of total glucan formation [12]. In addition, the inactivation of vicK produces a reduced level of lactic acid and enhances the acid tolerance of S. mutans [13]; a vicK knockout mutant has been shown to be more sensitive to H2O2 than the wild-type [8].
A vicR null mutation is apparently lethal to S. mutans. VicR acts directly on the promoter regions of the gtfB, gtfC, and ftf genes. Overproduction of the VicR transcript upregulates these genes [12]. In addition, VicR binds specifically to the comC gene, thereby negatively affecting the transcription of comC, comDE, comX, and nlmC [10]. Moreover, genes such as atlA, bmsH, glnQ, copy, wapA, relR, gcrR, plsX, and nlmC, demonstrate direct binding by S. mutans VicR. Among these genes, VicR negatively regulates nlmC and copY and positively regulates atlA, bmsH [14].
Early childhood caries (ECC) remains one of the most common chronic diseases of childhood [15, 16], particularly in China. Many reports have stated that S. mutans demonstrates a strong relationship with ECC [17, 18]. However, not every individual colonized by S. mutans will have dental decay. The DNA loci associated with strains isolated from children with ECC are different from those found in caries-free children [19, 20]. The VicRK system is important to S. mutans growth and virulence; therefore, the purpose of this study was to explore the genetic polymorphisms of the vicR and vicK genes found in S. mutans, which are associated with a distinct caries experience in children.
Methods
Bacterial strains
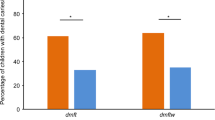
This study, which is a continuation of a previous study [21], primarily aimed to explore the connections between the missense mutations of vicR and vicK genes and the severity of caries in children with S. mutans. A case-control group study design was used, and the sample was the same as that used in the previous study [21], in which the sample capacities were set after calculation. All statistical tests were two-sided, and the value of α was set at 0.05. The study subjects were selected from our previous study, which was performed in the Huadu District of Guangzhou in South China. The study protocol was approved by the Ethics Committee of Guanghua School of Stomatology, Sun Yat-sen University (ERC-[2012]-13). The procedures used were previously described [21]. Briefly, 121 caries-free children who were S. mutans-positive were selected as the caries-free group using a simple random sampling method. Each S. mutans-positive sample was numbered in the caries-free group, and then 121 samples were selected according to the random numbers generated by the computer. To determine the between-group genetic differences in the vicR and vicK genes of S. mutans, children who were S. mutans-positive and had a distinct caries status (dmft ≥6) formed the high-severity caries group. The dmft score in the high-severity group was based on the category used in a previous study [22]. In this study, one isolated S. mutans strain was selected from each child.
Extraction of chromosomal DNA
Plaque samples from the children were mixed, sonicated and then dispersed to obtain 10− 3 dilutions. The S. mutans strains were identified according to the colony morphology, and the colonies were tested for their ability to ferment mannitol, sorbitol, raffinose, melibiose, and aesculin as well as for their ability to hydrolyse arginine. The S. mutans strains were cultured in 2 ml of brain-heart infusion broth and incubated under anaerobic conditions (85% N2, 5% CO2, and 10% H2) at 37 °C for 18 h. The bacterial cells were harvested by centrifugation at 5000 rpm (~ 2800×g) for 5 min. Cells were resuspended in 5% Chelex100 buffer (containing 1% Tween-20, 0.03% SDS and 1% NP40). To lyse S. mutans, the suspension was treated with 10 units of proteinase K (20 mg/ml) at 37 °C for 1 min. The mixture was incubated at 56 °C for 1 h, followed by boiling for 10 min and immediate cooling on ice for 3 min. The DNA sample was collected according to the protocol of TIANamp Bacteria DNA Kit (Tiangen, Beijing, China). The obtained samples were measured with a UV spectrophotometer at 260 nm and 280 nm to detect the quality and quantity of DNA.
Amplification of the vicR and vicK genes
S. mutans UA159 was used as a reference strain. The total length of the vicR gene was 708 bp (Gene ID: 1028759), and the vicK gene length was 1353 bp (Gene ID: 1028760). ABI Primer Designer V3.0 was used to design the PCR primers. The vicR gene was PCR-amplified using the paired primers 5’-CGGGATCCATGAAGAAAATTCTAATCGTTGACGA-3′ (BamHI site italic) and 5’-CCGCTCGAGTTAGTCATATGATTTCATGTAATAAC-3′ (XhoI site italic). The vicK gene was PCR-amplified using the paired primers 5’-CGGGATCCATGACTAATGTGTTTGAATCAAGTC-3′ (BamHI site italic) and 5’-CCGCTCGAGTCATGATTCGTCTTCATCTTCTTCC-3′ (XhoI site italic).
The PCR reaction was performed in a 25 μl reaction volume containing 2.5 μl of 10× PCR buffer, 1.5 mM of MgCl2, 0.2 mM of dNTP mix, 0.2 μM of each primer, 100–300 ng of genomic DNA as a template, and 2 U of Platinum® Taq DNA Polymerase (Invitrogen, Carlsbad, CA, USA). The PCR cycling profile included an initial preheating step at 95 °C for 5 min, followed by denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and elongation at 72 °C for 50 s. A total of 30 cycles were performed, followed by a final elongation step at 72 °C for 5 min. The PCR amplification product was analysed by electrophoresis in a 1.5% agarose gel.
Sequencing of the vicR and vicK genes
The PCR products were purified using a QIAquick Gel Extraction Kit (QIAgen, Hilden, Germany), according to the manufacturer’s protocol. Ultimately, sequencing was performed with a 3730XL DNA analyser platform (Shanghai Life Technologies Biotechnology Company, Life Technologies, Shanghai, China). The PCR product sequences were compared with known vicR and vicK gene sequences from the UA159 strain in GenBank using Variant Reporter software.
Statistical analysis
Data analysis was performed using the SPSS 20.0 software. A Chi-square test was used to analyse single nucleotide polymorphisms of the vicR and vicK genes in S. mutans in the high-severity caries group and the caries-free group. A P-value < 0.05 for the chi-squared test was considered statistically significant.
Results
All 242 clinically isolated S. mutans strains included the vicR and vicK genes. No nucleotide sequence insertions or deletions were observed in the two genes.
Amino acid transversion, according to the vicR gene codons, is shown in Table 1. Four silent point mutations were identified. No missense mutations were found. The distribution frequencies of the silent mutation sites of the vicR gene in the high-severity caries and caries-free groups are shown in Table 2.
Amino acid transversion, according to the vicK gene codons, is shown in Table 3. A total of 41 mutations were identified. In addition to the 32 silent point mutations, 9 point mutations were found, resulting in missense mutations at the 173, 337, 470, 1051, 1132, 1258, 1260, 1277, and 1348 bp positions. The distribution frequencies of the mutation sites of the vicK gene in the high-severity caries and caries-free groups are shown in Table 4. The distribution of the C470T missense mutations was higher in the high-severity caries group than in the caries-free group (P = 0.002), and the other missense mutation rates in the two groups did not demonstrate any statistically significant differences (Table 5). The C470T missense mutation denotes a C base at the 470th base in the vicK gene of S. mutans UA159, while a T base is substituted in the vicK gene of clinical isolates.
Discussion
The profound effects of genetic polymorphisms of virulence factors on S. mutans have been a focus of caries research for many years [23, 24]. The vicRK signal transduction system is essential for S. mutans by modulating gene expression. However, all prior studies have focused on constructing mutants of laboratory reference strains, and little is known about the gene variation in clinical isolates and its relationship with caries. Compared to laboratory reference strains, clinical isolates are closer to the true disease state.
Hence, in this study, we sequenced the vicR and vicK genes of S. mutans strains isolated from children with a distinct caries status to analyse the effects of vicR and vicK polymorphisms on the risk of ECC. Mutational analysis showed that VicR in S. mutans plays an essential role in the viability of this bacterium [12]. In our study, all 242 clinical isolates had a full-length vicR gene, and no missense mutations were found, which indicated that the vicR gene was highly conserved among the clinical isolates. This ubiquitous distribution and sequence conservation of vicR among the S. mutans clinical isolates may suggest that it plays an important role in S. mutans biology.
The overall structure of VicK is a long-rod dimer that anchors four connected domains: HAMP (aa 36–86), PAS (aa 87–198), DHp (aa 199–269) and CA (aa 278–450) [25]. Compared to S. mutans UA159, all 242 clinical isolates had a full-length vicK gene, and nine missense mutation sites were found. All missense mutation sites identified in the present study were distributed in the HAMP, PAS and CA domains. No missense mutation sites were found in the DHp domain. The statistical analysis of the missense mutations was worthy of attention because these changes could affect protein activity. The statistical analysis showed that the differences in missense mutation rates between the caries-free and high-severity caries groups were not significant, except for that of the C470T mutations. The frequency of the C470T missense mutation was higher in the high-severity caries group than in the caries-free group. The cytosine (C) to thymine (T) mutation at the 470th bp in the vicK gene leads to a mutation of threonine (T) to methionine (M) at the amino acid residue at position 157 in the VicK amino acid sequence. The 157-amino acid residue belongs to the PAS domain. The PAS domain is a major sensor, with adopted canonical folds and dyad symmetry. The effect of the mutation at site 470 (aa 157) on the activity and function of the VicK protein requires further study.
The present study has some limitations. First, a dmft level of ≥6 was set as the cut-off value, which may have narrowed the selection of candidates for the high-severity caries group. However, this experiment was exploratory in nature. Children with a distinct caries status were utilised to explore and compare the genetic polymorphisms in the vicR and vicK genes of S. mutans. Second, the sample size was relatively small; therefore, a larger sample size may be required for additional between-group analysis.
Conclusions
The present study provided knowledge of the genetic diversity of the vicR and vicK genes of S. mutans in children with two distinct caries experiences: caries-free and high-severity caries. The results indicated that in children, sequences of vicR are highly conserved in S. mutans clinical isolates and the C470T missense mutation of the vicK gene may be related to caries experience with S. mutans.
Abbreviations
- dmft:
-
Decayed, missing and filled teeth
- DNA:
-
Deoxyribonucleic acid
- ECC:
-
Early childhood caries
- PCR:
-
Polymerase chain reaction
- S. mutans:
-
Streptococcus mutans
- TCRSs:
-
Two-component regulatory systems
References
Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721–32.
Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–7.
Krzysciak W, Jurczak A, Koscielniak D, Bystrowska B, Skalniak A. The virulence of Streptococcus mutans and the ability to form biofilms. Eur J Clin Microbiol Infect Dis. 2014;33(4):499–515.
Nicolas GG, Lavoie MC. Streptococcus mutans and oral streptococci in dental plaque. Can J Microbiol. 2011;57(1):1–20.
West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26(6):369–76.
Wolanin PM, Thomason PA, Stock JB. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 3(10):2002, REVIEWS3013.
Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A. 2002;99(22):14434–9.
Deng DM, Liu MJ, Ten Cate JM, Crielaard W. The VicRK system of Streptococcus mutans responds to oxidative stress. J Dent Res. 2007;86(7):606–10.
Tremblay YDN, Lo H, Li YH, Halperin SA, Lee SF. Expression of the Streptococcus mutans essential two-component regulatory system VicRK is pH and growth-phase dependent and controlled by the LiaFSR three-component regulatory system. Microbiology. 2009;155(9):2856–65.
Senadheera DB, Cordova M, Ayala EA, Chavez De Paz LE, Singh K, Downey JS, Svensater G, Goodman SD, Cvitkovitch DG. Regulation of Bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J Bacteriol. 2012;194(6):1307–16.
Biller SJ, Wayne KJ, Winkler ME, Burkholder WF. The putative hydrolase YycJ (WalJ) affects the coordination of cell division with DNA replication in Bacillus subtilis and may play a conserved role in Cell Wall metabolism. J Bacteriol. 2011;193(4):896–908.
Senadheera MD, Guggenheim B, Spatafora GA, Huang YC, Choi J, Hung DC, Treglown JS, Goodman SD, Ellen RP, Cvitkovitch DG. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol. 2005;187(12):4064–76.
Senadheera D, Krastel K, Mair R, Persadmehr A, Abranches J, Burne RA, Cvitkovitch DG. Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J Bacteriol. 2009;191(20):6415–24.
Ayala E, Downey JS, Mashburn-Warren L, Senadheera DB, Cvitkovitch DG, Goodman SD. A biochemical characterization of the DNA binding activity of the response regulator VicR from Streptococcus mutans. PLoS One. 2014;9(9):e108027.
Parker EJ, Jamieson LM, Broughton J, Albino J, Lawrence HP, Roberts-Thomson K. The oral health of indigenous children: a review of four nations. J Paediatr Child Health. 2010;46(9):483–6.
Folayan MO, Chukwumah NM, Onyejaka N, Adeniyi AA, Olatosi OO. Appraisal of the national response to the caries epidemic in children in Nigeria. Bmc Oral Health. 2014;14:76.
Saraithong P, Pattanaporn K, Chen Z, Khongkhunthian S, Laohapensang P, Chhun N, Pattanaporn W, Gaw HY, Li Y. Streptococcus mutans and Streptococcus sobrinus colonization and caries experience in 3- and 5-year-old Thai children. Clin Oral Investig. 2015;19(8):1955–64.
Parisotto TM, Steiner-Oliveira C, Silva CM, Rodrigues LK, Nobre-dos-Santos M. Early childhood caries and mutans streptococci: a systematic review. Oral Health Prev Dent. 2010;8(1):59–70.
Saxena D, Caufield PW, Li Y, Brown S, Song J, Norman R. Genetic classification of severe early childhood caries by use of subtracted DNA fragments from Streptococcus mutans. J Clin Microbiol. 2008;46(9):2868–73.
Zhou Q, Qin X, Qin M, Ge L. Genotypic diversity of Streptococcus mutans and Streptococcus sobrinus in 3-4-year-old children with severe caries or without caries. Int J Paediatr Dent. 2011;21(6):422–31.
Yu LX, Tao Y, Qiu RM, Zhou Y, Zhi QH, Lin HC. Genetic polymorphisms of the sortase a gene and social-behavioural factors associated with caries in children: a case–control study. Bmc Oral Health. 2015;15(1)
Martins-Junior PA, Vieira-Andrade RG, Correa-Faria P, Oliveira-Ferreira F, Marques LS, Ramos-Jorge ML. Impact of early childhood caries on the oral health-related quality of life of preschool children and their parents. Caries Res. 2013;47(3):211–8.
Napimoga MH, Hofling JF, Klein MI, Kamiya RU, Goncalves RB. Tansmission, diversity and virulence factors of Sreptococcus mutans genotypes. J Oral Sci. 2005;47(2):59–64.
Lemos JA, Quivey RJ, Koo H, Abranches J. Streptococcus mutans: a new gram-positive paradigm? Microbiology. 2013;159(Pt 3):436–45.
Wang C, Sang J, Wang J, Su M, Downey JS, Wu Q, Wang S, Cai Y, Xu X, Wu J, et al. Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains. PLoS Biol. 2013;11(2):e1001493.
Acknowledgements
We are grateful for the support of participants of the research group.
Funding
This work was supported by Grant No. A2015419 from Medical Scientific Research Foundation of Guangdong Province, and Grant No. 2016A020215067 and No. 2014A020212092 from Guangdong Provincial Science and Technology Plan Project, China. The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets generated and/or analysed during the current study are not publicly available due to anonymity considerations but are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
PLZ and LXY contributed to the study design, the experiment conduct and manuscript preparation. JKL and YZ analysed and interpreted focus data. HCL contributed to the study conception and design, general research group supervision, and critical manuscript revision. All authors have read and approved the final version of this manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Human subject approval was received from the Ethical Review Committee of Guanghua School of Stomatology, Hospital of Stomatology, Institute of Stomatological Research, Sun Yat-Sen University. Written informed consent was given by all focus group participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhuang, P.L., Yu, L.X., Liao, J.K. et al. Relationship between the genetic polymorphisms of vicR and vicK Streptococcus mutans genes and early childhood caries in two-year-old children. BMC Oral Health 18, 39 (2018). https://doi.org/10.1186/s12903-018-0501-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12903-018-0501-y