Abstract
Background
The production of chemokines by tissue resident cells during inflammation is considered one of the main mechanisms involved in the formation of inflammatory infiltrates. Fibroblasts are the main resident cell type in gingival and periodontal ligament tissues, and their ability to produce chemokine stromal cell-derived factor 1 alpha (SDF-1α) and its receptor CXCR4 under stimulation by gram negative bacteria, Porphyromonas gingivalis, commonly found in periodontal infections was investigated.
Methods
Western blots were used to assess SDF-1α and CXCR4 protein expression levels in human gingival fibroblast cells (HGF-1) induced by Lipopolysaccharide (LPS) from P. gingivalis in the presence or absence of LY294002, a highly selective inhibitor of PI-3K/Akt. RT-PCR and quantitative Real-time PCR was performed using gingival mRNAs from periodontitis patients. Immunohistochemistry was performed to analyze the expression and subcellular localization of SDF-1α and CXCR4, together with NF-kβ phosphorylation, in specimens from patients with periodontitis and in an experimental rat periodontitis model.
Results
We found that P. gingivalis LPS up-regulated SDF-1α and CXCR4 protein levels and elevated phosphorylation of the SDF-1α-responsive NF-kβ and Akt at 24 h in HGF-1 cells. SDF-1α and CXCR4 mRNA and protein expression levels were high in all patients with periodontitis. In the P. gingivalis-induced rat experimental periodontitis model, SDF-1α and CXCR4 immunoreactivity was higher in gingival and periodontal ligament tissues compared to the control.
Conclusion
Our data showed that PI-3K/Akt is an upstream participant in the P. gingivalis LPS-mediated induction of SDF-1α. Taken together, these results suggest that the chemokine SDF-1α and its receptor CXCR4 contribute to P. gingivalis-induced periodontal inflammation.
Similar content being viewed by others
Background
Fibroblasts are tissue-resident cells presenting unique phenotypes according to their tissue of origin. Recently, fibroblasts have been suggested to be important sentinel cells in the immune system [1] and their reactions to various microbial stimuli, including Gram-negative bacteria-derived virulence factors, have been studied in diverse situations [2–4]. Fibroblasts actively define the structure of the tissue microenvironment and regulate inflammatory responses by producing cytokines and chemokines [1, 5]. Lipopolysaccharide (LPS) derived from the outer membrane of the Gram-negative bacteria, Porphyromonas gingivalis (P. gingivalis), is one of the most commonly investigated virulence factors used to activate the innate immune system [6, 7].
Periodontal pathogens, including P. gingivalis, can cause inflammation [8, 9], which results in soft tissue destruction and periodontal bone resorption in the development of periodontal disease. The host-immune response to bacteria has been suggested to be associated with the alteration or even the progression of this disease [10, 11]. A wide variety of cytokines, chemokines and their receptors are synthesized by gingival fibroblasts, epithelial cells, endothelial cells and inflammatory cells [12, 13]. The production of chemokines by those cells may vary significantly when comparing fibroblasts from different tissues [14–16], thus suggesting their important role in inflammatory infiltrate formation and cellular traffic during tissue repair.
The chemokine CXCL12, also known as stromal cell-derived factor 1 alpha (SDF-1α), is constitutively expressed by human gingival fibroblasts (HGFs) and by human periodontal ligament (PDL) fibroblasts (HPDLFs). CXCL12 is responsible for regulating the trafficking of bone marrow progenitor cells, as well as the transendothelial migration of leukocytes [17, 18]. SDF-1α plays a role as an essential and non-redundant factor involved in tissue remodeling, specifically in vascular regeneration [19]. Increased SDF-1α gene expression has also been reported in an experimental model of bacterial-induced apical periodontitis [20, 21]. Individuals with periodontal disease have higher levels of SDF-1α in their gingival crevicular fluid compared to healthy individuals and neutrophil migration is also enhanced in the presence of this chemokine [22]. In addition, SDF-1α from PDL cells was demonstrated to participate in the regeneration and homeostasis of periodontal tissues via the recruitment of stem cells [23]. SDF-1α has been shown to promote cell viability and proliferation, to affect differentiation and to exert a migratory effect on PDL stem cells in vitro [24]. Moreover, SDF-1α is up-regulated after myocardial infarction, bone or cartilage fracture, and ischemic cerebral injury at injured sites [25]. Nevertheless, the involvement of SDF-1α in the progression of periodontal inflammation and its potential interactions with signaling cascades are not yet known.
SDF-1α regulates numerous homeostatic and pathological processes through its receptor CXCR4, by inducing several signaling transduction pathways, including activation of the PI-3K/Akt-NF-kβ axes [26, 27]. Expression of CXCR4 is regulated by NF-kβ and the CXCR4 promoter contains p50/p65 binding sites [26, 28]. Moreover, stimulation of mesoangioblasts with SDF-1α triggers the nuclear translocation of NF-kβ p65, which is required for mesoangioblast migration in response to SDF-1α [29].
Considering the complex reciprocal functional interactions that the pleiotropic chemokine SDF-1α can establish with the periodontal microenvironment, the aim of this study was to evaluate SDF-1α and CXCR4 in fibroblasts of gingiva and of PDL origin in terms of the signaling cascade of PI-3K/Akt and the regulation of NF-kβ. Human gingival fibroblast cells stimulated with LPS from P. gingivalis exhibited an increased expression of SDF-1α and CXCR4 via activation of the PI3K/Akt and NF-kβ signaling pathways. By employing the P. gingivalis-induced experimental rat periodontitis model, we provide evidence that SDF-1α and CXCR4 proteins play an inflammation-promoting role in the development of periodontitis by regulating the function of NF-kβ.
Methods
Cell culture
HGF-1 cells were obtained from the American Type Culture Collection (CRL-2014, ATCC, Manassas, VA, USA) and were cultured in Dulbecco’s modified Eagles medium (DMEM) containing 10 % fetal bovine serum (FBS). HGF-1 cells were seeded in 60-mm plastic tissue culture dishes and incubated in 5 % CO2 at 37 °C. When the cells reached sub-confluence, they were harvested and sub-cultured. The cells at the fourth passage were used in the experiments. LPS from P. gingivalis was added to the cultures for 24 h to evaluate the effects of treatment with an inflammatory cytokine. Where noted, cells were treated with 10 μM LY294002 (Calbiochem, San Diego, CA. USA) for 1 h prior to LPS treatment. The concentration of LY294002 was adopted from our previous study [30].
Preparation of bacteria
P. gingivalis ATCC 33277 was grown in brain heart infusion broth supplemented with 5 mg/ml yeast extract, 5 μg/ml hemin and 0.2 μg/ml vitamin K1, as described previously [31]. Bacterial cells were grown under anaerobic conditions (85 % N2, 10 % H2 and 5 % CO2) at 37 °C for 24 h. LPS from P. gingivalis ATCC 33277 was obtained from P. gingivalis according to the manufacturers’ instructions (iNtRON Biotechnology, Kyungki-Do, Korea). In brief, 5 ml of a bacterial cell suspension were centrifuged for 30 s at 13,000 rpm at room temperature to remove all traces of the supernatant, and were then vortexed vigorously with 1 ml lysis buffer. Two hundred μl chloroform were then added and vortexed vigorously for 20 s, and then incubated at room temperature for 5 min. The suspension was then centrifuged at 13,000 rpm for 10 min at 4 °C and then 400 μl of the supernatant was transferred to a new 1.5 ml tube, mixed with 800 μl purification buffer and incubated for 10 min at −20 °C. The sample was then centrifuged at 13,000 rpm for 15 min at 4 °C. After washing the LPS pellet with 1 ml 70 % EtOH, it was dried completely. Seventy μl of 10 mM Tris-HCl buffer (pH 8.0) were added to the LPS pellet and sonicated.
Experimental periodontitis
An established method of experimental periodontitis has been previously reported [31]. Briefly, twelve 5 week-old male Sprague-Dawley rats (CLEA Japan, Inc., Tokyo, Japan) were given sulfamethoxazole (1 mg/ml) and trimethoprim (200 μg/ml) in their drinking water for 4 days to reduce any existing oral microorganisms, followed by a 3 day antibiotic-free period before starting the oral challenges with bacteria. Rats had free access to laboratory chow and tap water. They were randomly divided into two experimental groups [Group A: 5 % carboxymethylcellulose (CMC) (control group); Group B: P. gingivalis ATCC 33277 (P. g. group)] of 6 rats each. Each rat infected with P. gingivalis received 0.5 ml (1.0 × 108 cells/ml) of the bacterial suspension in 5 % CMC by oral gavage at 8, 10 and 12 days. All rats were sacrificed by CO2 inhalation 30 days after the last gavage.
Immunohistochemistry
In total, 17 formalin-fixed, paraffin-embedded gingival tissues were obtained from chronic periodontitis patients (9 male, 8 female). Gingival tissues were obtained from the individuals undergoing periodontal surgery, showed moderate to severe disease and were classified as a periodontitis group with probing depths >4-6 mm. The diagnostic criteria for periodontal disease were performed according to the American Academy of Periodontology [32]. Plaque index (PLI) and gingival index (GI) were evaluated as proposed by Silness and Loe’s method [33]. Clinical attachment level (CAL) was determined by measuring the distance from the periodontal pocket depth to the cementoenamel junction. Severity is based on the amount of CAL and is designated as moderate (4 mm CAL) or severe (>5 mm CAL). Clinical measurements, including probing of pocket depth (PD), bleeding on probing (BOP) were recorded using a manual probe (PCP-UNC 15, Hu-Friedy Chicago, IL, USA) at six sites per tooth and the reading was recorded to the nearest 1 mm. Formalin-fixed, paraffin-embedded gingival tissue sections were immunostained for SDF-1α, CXCR4, and phospho NF-kβ, with the CSA II System (Dako, Carpinteria, CA, USA), in accordance with the manufacturer’s instructions. Sections were initially immersed in Target Retrieval Solution (DAKO) at 95 °C for 12 min, and then cooled for 30 min. Endogenous peroxidase activity was blocked with REAL Peroxidase-Blocking Solution (S2023, DAKO) for 30 min. Antibodies against SDF-1α (1:50; Abcam, Cambridge, MA, USA), CXCR4 (1:50; Abcam) and phospho NF-kβ (1:100; Bioss, Inc., Woburn, MA, USA), were used as primary antibodies and were incubated overnight at 4 °C. The secondary antibodies conjugated to peroxidase (Nichirei Biosciences, Tokyo, Japan) were incubated at room temperature for 30 min. After rinsing with PBS, all specimens were color developed with a 3,3′-Diaminobenzidine tetrahydrochloride (DAB) chromogen kit (Dako), counterstained with hematoxylin, and examined by light microscopy. The immunostaining of all specimens was performed simultaneously to ensure the same antibody reaction and DAB exposure conditions.
RNA extraction, RT-PCR and quantitative RT-PCR analysis
Total RNA was extracted from gingival tissues using an RNeasy Mini Kit (Qiagen, Tokyo, Japan) and reverse-transcribed using High Capacity RNA-to-cDNA Master Mix (Applied Biosystems, Foster City, CA, USA). For RT-PCR, the reaction mixture (20 μL) contained 1 μL of diluted cDNA sample and 10 pmol of each pair of oligonucleotide primers. PCR conditions included an initial denaturation at 95 °C for 10 min, followed by a 30-cycle amplification consisting of denaturation at 94 °C for 15 s, annealing at 55 °C for 30 s and extension at 72 °C for 30 s. The primers used in RT-PCR analysis are as follows: SDF-1 (5′-agagccaacgtcaagcatct-3′ Forward, 5′-gggcagcctttctcttcttc-3′ Reverse); CXCR4 (5′-ctgagaagcatgacggacaa-3′ Forward, 5′-tcgatgctgatcccaatgta-3′ Reverse); β-actin (5′-agccatgtacgttgcta-3′ Forward, 5′-agtccgcctagaagca-3′ Reverse). All primer pairs were checked for primer-dimer formation using the three-step protocol described above without the addition of the RNA template. For the standard PCR, the products were separated on 1.5 % agarose gels and visualized by ethidium bromide staining. The relative expression levels of target mRNAs, compared to the level of β-actin RNA, were analyzed by real time PCR with the corresponding TaqMan MGB probes (Hs03676656_mH for SDF-1α, Hs00607978_s1 for CXCR4, and Hs99999903_m1 for β-actin) using QuantStudio 6 Real Time PCR System (Applied Biosystems). The thermal cycling conditions were according to the TaqMan Fast Universal PCR protocol.
Western blot analysis
HGF-1 cells were harvested in RIPA lysis buffer (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Samples were boiled for 5 min, chilled on ice for 5 min, and centrifuged. Equal amounts of protein (20 μg) were electrophoresed on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels and electrophoretically transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA, USA). The membranes were blocked with the blocking solution in TBS containing 0.01 % Tween 20 to reduce nonspecific binding, probed overnight at 4 °C with the following primary antibodies: anti-SDF-1 rabbit polyclonal antibody (1:500, Abcam, Cambridge, MA, USA), anti-CXCR4 rabbit monoclonal antibody (1:500, Abcam), anti-Akt rabbit monoclonal antibody (1:1000; Cell Signaling Technology, Beverly, MA, USA), anti-phospho Akt rabbit monoclonal antibody (1:2000; Cell Signaling Technology), anti-NF-kβ p65 rabbit monoclonal antibody (1:1000; Cell Signaling Technology), and anti-phospho NF-kβ p65 rabbit polyclonal antibody (1:1000; Cell Signaling Technology). Anti-β-actin rabbit polyclonal antibody (1:1000; Cell Signaling Technology) was used for loading control. The blots were then incubated for 1 h with a horseradish peroxidase–conjugated anti-rabbit secondary antibody (1:2000; Cell Signaling Technology). Protein bands were developed by ECL plus Western blotting detection reagent (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) and imaged with an ImageQuant LAS 4000 Mini (GE Healthcare Bio-Sciences). Protein bands were scanned and analyzed by densitometry using the ImageJ software (NIH).
Statistical analysis
Significant differences were analyzed by Fisher’s exact test. A P-value of less than 0.05 is considered statistically significant.
Results
Effects of P. gingivalis LPS on the expression of SDF-1α and CXCR4 in HGF-1 cells
To understand whether inflammatory gingival activation alters the expression levels of SDF-1α and/or CXCR4, cultures of HGF-1 cells were exposed to P. gingivalis LPS at different concentrations, and SDF-1α and CXCR4 protein expression levels were evaluated by western blotting. In line with the cytokine functions of SDF-1α, the data demonstrate that P. gingivalis LPS increased SDF-1α and CXCR4 expression levels after 24 h of exposure (Fig. 1a). Moreover, the data in Fig. 1a show that P. gingivalis LPS stimulated NF-kβ and Akt phosphorylation in a dose-dependent manner.
a Effect of LPS on the expression of SDF-1α and CXCR4 in HGF-1 cells. HGF-1 cells were incubated with different concentrations of LPS from P. gingivalis for 24 h. The cell lysates were then assayed to determine the expression of SDF-1α and CXCR4, Akt and NF-kβ p65, and the phosphorylation of Akt and NF-kβ p65 using Western blots. Membranes were stripped and re-probed with an anti-β-actin antibody as a loading control. Protein bands were quantified by densitometric analyses. The results are expressed as means ± S.D. (*p < 0.05). b Role of PI-3K/Akt in P. gingivalis LPS-stimulated SDF-1α expression. HGF-1 cells were pre-treated with or without LY294002 for 1 h and were then incubated with or without P. gingivalis LPS (500 μM) for 24 h. Cell lysates were assayed using Western blots to determine the expression of SDF-1α. Membranes were stripped and re-probed with an anti-β-actin antibody as a loading control. Protein bands were quantified by densitometric analyses. The results are expressed as means ± S.D. (*p < 0.05)
Involvement of PI3-K/Akt in P. gingivalis LPS-induced expression of SDF-1α
To determine whether the PI-3K/Akt cascade plays an important role in the LPS-induced expression of SDF-1α in HGF-1 cells, pre-treatment of HGF-1 cells with a pharmacological inhibitor of PI-3K, LY294002, significantly attenuated the P. gingivalis LPS-stimulated expression of SDF-1α, suggesting the involvement of PI-3K/Akt in that increased expression (Fig. 1b). The concentration of P. gingivalis LPS used (500 μM) was adopted from the results shown in Fig. 1a. A preliminary screening was done to obtain the optimal concentration of LY294002 using the MTS assay and the LDH cytotoxicity assay with P. gingivalis LPS-treated cells (data not shown). These results suggest that PI-3K/Akt plays an important role in the P. gingivalis LPS-induced expression of SDF-1α in HGF-1 cells.
Periodontitis stimulates SDF-1α and CXCR4 mRNA in human periodontal tissues
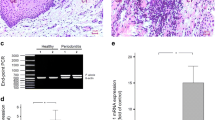
In order to confirm the expression levels of SDF-1α and CXCR4 in human periodontal tissues, their expression was evaluated using RT-PCR and quantitative real-time PCR. HGF-1 cells were used as a positive control. SDF-1α and CXCR4 mRNAs were identified in HGF-1 cells and in tissue homogenates from human periodontal tissues (Fig. 2).
Level of SDF-1α and CXCR4 mRNAs in human periodontal tissues. Total RNA was isolated from each sample and was subjected to RT-PCR and quantitative real-time PCR analysis. SDF-1α and CXCR4 mRNAs were highly expressed in human periodontitis tissues. Relative mRNA levels were calculated as a ratio to the housekeeping gene (β-actin). Each bar represents the mean ± SD for at least 3 independent experiments. HGF-1 cells were used as a positive control
Increased SDF-1α and CXCR4 immunostaining in human periodontal tissues
In chronic inflammatory conditions, SDF-1α and CXCR4 expression was observed in suprabasal layers of the epithelium of periodontitis patients. There was also positive staining for SDF-1α and CXCR4 present in some nuclei in the basal layer (Fig. 3). Faint expression of SDF-1α and CXCR4 protein was found in the epithelium of nonperiodontitis controls.
Immunohistochemical analysis of SDF-1α and CXCR4 in periodontal inflammation. Four μm thick sections of formalin-fixed, paraffin-embedded specimens were deparaffinized and immunoreactivity was detected using a DAKO ENVISION Kit. SDF-1α and CXCR4 was highly expressed in patients with periodontitis. SDF-1α and CXCR4 expression in human periodontal tissues was present predominantly in the granular and spinous layers of epithelial cells, while their expression was faint in nonperiodontitis tissues. SDF-1α and CXCR4 expression was also present in the suprabasal layer of epithelial cells. Scale bars, 50 μM
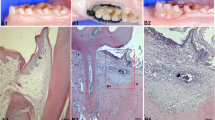
P. gingivalis-induced gingival inflammation triggers activation of the SDF-1α/NF-kβ signaling pathway
SDF-1α and CXCR4 immunohistochemistry revealed a strong staining in P. gingivalis challenged gingival epithelium compared to the control (Fig. 4a). SDF-1α and CXCR4 were only weakly expressed in the gingiva of control animals. A strong reaction with anti-SDF-1α and anti-CXCR4 antibodies was found in P. gingivalis challenged PDL cells (Fig. 4b). Analysis of the immunostaining pattern of NF-kβ phosphorylation in the experimental rat periodontitis model revealed that it was mainly localized to inflammatory infiltrates in PDL cells (Fig. 4b), whereas no detection was observed in control PDL cells.
Immunohistochemical analysis of SDF-1α and CXCR4 in P. gingivalis challenged experimental rat periodontal inflammation. SDF-1α and CXCR4 were abundantly expressed in the P. gingivalis challenged rat tissues. Immunohistochemical analysis revealed a higher expression of SDF-1α and CXCR4 in P. gingivalis challenged rat gingiva (Fig. 4a; Scale bars, 20 μM) and PDL (Fig. 4b; Scale bars, 100 μM) compared to the control
Discussion
In this study, an up-regulation in SDF-1α protein expression by HGF-1 cells was detected in a concentration-dependent manner when they were stimulated with LPS from P. gingivalis. The induction of phospho NF-kβ protein expression when cells were challenged by LPS was significant for HGF-1 cells in vitro and PDL cells in vivo, highlighting the important participation of this molecule in inflammatory events of those tissues. We detected an increase in SDF-1α and CXCR4 expression by HGF-1 cells starting at 6 h (data not shown), which was maintained at 24 h. We observed a trend of increased expression depending on the LPS concentration in HGF-1 cells. Moreover, P. gingivalis is an important factor of SDF-1α/CXCR4 expression by HGFs in periodontal tissue, and the increase of SDF-1α and CXCR4 expression might be related to the progression of periodontal disease. Taking this into account, it is possible that the induction of SDF-1α expression contributes to the inflammation because, as documented in this study, the expression of signaling molecules, such as NF-kβ, seems to occur. Another study [22] demonstrated that SDF-1α levels were altered due to periodontal inflammation, thus corroborating our speculation regarding the participation of SDF-1α as an important chemokine in the inflammatory microenvironment context. The unveiling of the underlying molecular pathways regulating the inflammatory destruction of periodontal tissue during the progression of periodontitis could provide new and refined therapeutic approaches to prevent tooth loss.
The magnitude of HGF responses to LPS from P. gingivalis varied markedly between individuals, and in particular, it is possible that the heterogeneity in these responses was caused by the variability in the host’s genetic background of these cells [34]. Fedyk et al. [35] and Hosokawa et al. [7] demonstrated a decrease in SDF-1α in primary gingival fibroblasts under proinflammatory conditions. In contrast, Jiang et al. [36] showed the up-regulation of SDF-1α and its receptor CXCR4 in inflamed dental pulps. CXCR4 gene expression in periapical tissues was shown to be up-regulated after endodontic intervention in necrotic teeth [37], suggesting an important role of SDF-1α in tissue repair. On the basis of these data, it seems reasonable to assume that SDF-1α expression by fibroblasts is directly dependent on the tissue origin and bacterial product nature.
Considering that SDF-1α is an important chemoattractant for hematopoietic and mesenchymal stem cells supporting their survival and proliferation [38], and that this mediator exerts a migratory effect on PDL stem cells in vitro [24], it seems important to know that by knocking down Akt it would be possible to decrease crucial proinflammatory mediators, affecting chemokines involved in the repair process, such as SDF-1α. A PI-3K inhibitor, LY2940002, results in the inhibition of SDF-1α protein expression in this study. The current results indicate that the PI-3K/Akt pathway may relate to the LPS-induced expression of SDF-1α in HGF-1 cells. Signaling through the innate immune system by fibroblasts, the most numerous resident non-professional immune cells in the periodontium, controls the secretion of important cytokines that modulate the inflammatory microenvironment.
SDF-1α/CXCR4 signaling may have a dual role in inflammation and tissue repair. Participants with periodontal diseases have higher levels of SDF-1α and CXCR4 compared to healthy participants [22], and SDF-1α may recruit host-defensive cells as well as PDL cells to sites of inflammation to be involved in immune surveillance, wound healing and tissue repair. Similarly, expression of both SDF-1α and its receptor CXCR4 has been identified in human dental pulp cells and in stem cells, where those factors have been proposed to participate in the recruitment of cells to the sites of injury [36, 39]. PDL cells are thought to be responsible for the homeostasis and regeneration of periodontal tissues [40–42] following injury or inflammation by bacteria. Therefore, animal studies should be performed to elucidate the in vivo effects of SDF-1α on human PDL cells as well as on inflammatory cells. Our in vivo study detected abundant intracellular SDF-1α and its receptor CXCR4, which was also present on the cell membrane in a small fraction of PDL cells.
The up-regulated expression of SDF-1α at inflammation sites serves as a potent chemoattractant to recruit circulating or residing CXCR4-expressing cells to lesions, with high differentiation and proliferation potential to act as candidates to repair and regenerate the damaged tissues. The interaction of SDF-1α/CXCR4 may play an essential role in promoting the migration of CXCR4-expressing cells into damaged sites during tissue repair. The distinct ability of gingival and PDL fibroblasts to secrete SDF-1α and CXCR4 emphasizes that these cells may similarly contribute to the balance of cytokines in the LPS-challenged periodontium. Considering the importance of resident cells in leukocyte recruitment during local inflammation [43], understanding cellular protection mechanisms against bacterial products may significantly enhance our knowledge about the establishment of periodontal diseases. Our results suggest a potential strategy for in vivo therapies using SDF-1α to promote periodontal regeneration.
Conclusions
In conclusion, our results demonstrate that levels of SDF-1α and CXCR4 are up-regulated in periodontal inflammation. Since LPS from P. gingivalis stimulation increased NF-kβ and Akt activity, they may exert an influence on the levels of SDF-1α and CXCR4. Fibroblasts represent the major SDF-1α expressing cells, which suggest that this chemokine play an important role for sustained immune cell infiltration in periodontitis, particularly of fibroblasts.
Abbreviations
BOP, bleeding on probing; CAL, clinical attachment level; CMC, carboxymethylcellulose; DAB, 3,3′-Diaminobenzidine tetrahydrochloride; DMEM, Dulbecco’s modified Eagles medium; FBS, fetal bovine serum; GI, gingival index; HGFs, human gingival fibroblasts; HPDLFs, human periodontal ligament fibroblasts; LPS, Lipopolysaccharide; P. gingivalis, Porphyromonas gingivalis; PD, probing of pocket depth; PDL, human periodontal ligament; PLI, plaque index; SDF-1α, stromal cell-derived factor 1 alpha; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis
References
Pablos JL, Santiago B, Galindo M, Torres C, Brehmer MT, Blanco FJ, et al. Synoviocyte-derived CXCL12 is displayed on endothelium and induces angiogenesis in rheumatoid arthritis. J Immunol. 2003;170:2147–52.
Yamaji Y, Kubota T, Sasaguri K, Sato S, Suzuki Y, Kumada H, et al. Inflammatory cytokine gene expression in human periodontal ligament fibroblasts stimulated with bacterial lipopolysaccharides. Infect Immun. 1995;63:3576–81.
Wang PL, Oido-Mori M, Fujii T, Kowashi Y, Kikuchi M, Suetsugu Y, et al. Heterogeneous expression of Toll-like receptor 4 and downregulation of Toll-like receptor 4 expression on human gingival fibroblasts by Porphyromonas gingivalis lipopolysaccharide. Biochem Biophys Res Commun. 2001;288:863–7.
Wang PL, Azuma Y, Shinohara M, Ohura K. Toll-like receptor 4-mediated signal pathway induced by Porphyromonas gingivalis lipopolysaccharide in human gingival fibroblasts. Biochem Biophys Res Commun. 2000;273:1161–7.
Bradfield PF, Amft N, Vernon-Wilson E, Exley AE, Parsonage G, Rainger GE, et al. Rheumatoid fibroblast- like synoviocytes overexpress the chemokine stromal cell derived factor 1 (CXCL12), which supports distinct patterns and rates of CD4+ and CD8+ T cell migration within synovial tissue. Arthritis Rheum. 2003;48:2472–82.
Gutie’rrez-Venegas G, Maldonado-Frı’as S, Ontiveros-Granados A, Kawasaki-Ca’rdenas P. Role of p38 in nitric oxide synthase and cyclooxygenase expression, and nitric oxide and PGE2 synthesis in human gingival fibroblasts stimulated with lipopolysaccharides. Life Sci. 2005;77:60–73.
Hosokawa Y, Hosokawa I, Ozaki K, Nakae H, Murakami K, Miyake Y, et al. CXCL12 and CXCR4 expression by human gingival fibroblasts in periodontal disease. Clin Exp Immunol. 2005;141:467–74.
Dzink JL, Tanner AC, Haffajee AD, Socransky SS. Gram negative species associated with active destructive periodontal lesions. J Clin Periodontol. 1985;12:648–59.
Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent Jr RL. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44.
Kjeldsen M, Holmstrup P, Bendtzen K. Marginal periodontitis and cytokines: a review of the literature. J Periodontol. 1993;64:1013–22.
Wilson M, Reddi K, Henderson B. Cytokine-inducing components of periodontopathogenic bacteria. J Periodontal Res. 1996;31:393–407.
Okada H, Murakami S. Cytokine expression in periodontal health and disease. Crit Rev Oral Biol Med. 1998;9:248–66.
Graves DT. The potential role of chemokines and inflammatory cytokines in periodontal disease progression. Clin Infect Dis. 1999;28:482–90.
Hatakeyama J, Tamai R, Sugiyama A, Akashi S, Sugawara S, Takada H. Contrasting responses of human gingival and periodontal ligament fibroblasts to bacterial cell-surface components through the CD14/Toll-like receptor system. Oral Microbiol Immunol. 2003;18:14–23.
Morandini AC, Sipert CR, Gasparoto TH, Greghi SL, Passanezi E, Rezende ML, et al. Differential production of macrophage inflammatory protein-1alpha, stromal-derived factor-1, and IL-6 by human cultured periodontal ligament and gingival fibroblasts challenged with lipopolysaccharide from P. gingivalis. J Periodontol. 2010;81:310–7.
Scheres N, Laine ML, de Vries TJ, Everts V, van Winkelhoff AJ. Gingival and periodontal ligament fibroblasts differ in their inflammatory response to viable Porphyromonas gingivalis. J Periodontal Res. 2010;45:262–70.
Peled A, Grabovsky V, Habler L, Sandbank J, Arenzana-Seisdedos F, Petit I, et al. The chemokine SDF-1 stimulates integrin-mediated arrest of CD34 (+) cells on vascular endothelium under shear flow. J Clin Invest. 1999;104:1199–211.
Scimone ML, Felbinger TW, Mazo IB, Stein JV, Von Andrian UH, Weninger W. CXCL12 mediates CCR7-independent homing of central memory cells, but not naive T cells, in peripheral lymph nodes. J Exp Med. 2004;199:1113–20.
Ceradini DJ, Kulkarni AR, Callaghan MJ, Tepper OM, Bastidas N, Kleinman ME, et al. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med. 2004;10:858–64.
Fukada SY, Silva TA, Garlet GP, Rosa AL, da Silva JS, Cunha FQ. Factors involved in the T helper type 1 and type 2 cell commitment and osteoclast regulation in inflammatory apical diseases. Oral Microbiol Immunol. 2009;24:25–31.
Fukada SY, Silva TA, Saconato IF, Garlet GP, Avila-Campos MJ, Silva JS, et al. iNOS-derived nitric oxide modulates infection-stimulated bone loss. J Dent Res. 2008;87:1155–9.
Havens AM, Chiu E, Taba M, Wang J, Shiozawa Y, Jung Y, et al. Stromal-derived factor-1alpha (CXCL12) levels increase in periodontal disease. J Periodontol. 2008;79:845–53.
Asakawa T, Chosa N, Yoshimura Y, Asakawa A, Tanaka M, Ishisaki A, et al. Fibroblast growth factor 2 inhibits the expression of stromal cell derived factor 1a in periodontal ligament cells derived from human permanent teeth in vitro. Int J Mol Med. 2012;29:569–73.
Du L, Yang P, Ge S. Stromal cell-derived factor-1 significantly induces proliferation, migration, and collagen type I expression in a human periodontal ligament stem cell subpopulation. J Periodontol. 2012;83:379–88.
Lataillade JJ, Clay D, Dupuy C, Rigal S, Jasmin C, Bourin P, et al. Chemokine SDF-1 enhances circulating CD34(+) cell proliferation in synergy with cytokines: Possible role in progenitor survival. Blood. 2000;95:756–68.
Helbig G, Christopherson KW, Bhat-Nakshatri P, Kumar S, Kishimoto H, Miller KD, et al. NF-{kappa} B promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J Biol Chem. 2003;278:21631–8.
Neuhaus T, Stier S, Totzke G, Gruenewald E, Fronhoffs S, Sachinidis A, et al. Stromal cell-derived factor 1α (SDF-1α) induces gene-expression of early growth response-1 (Egr-1) and VEGF in human arterial endothelial cells and enhances VEGF induced cell proliferation. Cell Prolif. 2003;36:75–86.
Kukreja P, Abdel-Mageed AB, Mondal D, Liu K, Agrawal KC. Up-regulation of CXCR4 expression in PC-3 cells by stromal-derived factor-1alpha (CXCL12) increases endothelial adhesion and transendothelial migration: role of MEK/ERK signaling pathway-dependent NF-kappaB activation. Cancer Res. 2005;65:9891–8.
Palumbo R, Galvez BG, Pusterla T, De Marchis F, Cossu G, Marcu KB, et al. Cells migrating to sites of tissue damage in response to the danger signal HMGB1 require NF-{kappa} B activation. J Cell Biol. 2007;179:33–40.
Bhawal UK, Ito Y, Tanimoto K, Sato F, Fujimoto K, Kawamoto T, et al. IL-1β mediated up-regulation of DEC1 in human gingiva cells via the Akt pathway. J Cell Biochem. 2012;113:3246–53.
Bhawal UK, Lee HJ, Arikawa K, Shimosaka M, Suzuki M, Toyama T, et al. Micromolar sodium fluoride mediates anti-osteoclastogenesis in Porphyromonas gingivalis-induced alveolar bone loss. Int J Oral Sci. 2015;7:242–9.
Armitage GC. Research, Science and Therapy Committee of the American Academy of Periodontology. Diagnosis of periodontal diseases. J Periodontol. 2003;74:1237–47.
Silness J, Löe H. Periodontal disease in pregnancy. II. Correlation between oral hygiene and periodontal condition. Acta Odontol Scand. 1964;22:121–35.
Souza PP, Palmqvist P, Lundgren I, Lie A, Costa-Neto CM, Lundberg P, et al. Stimulation of IL-6 cytokines in fibroblasts by toll-like receptors 2. J Dent Res. 2010;89:802–7.
Fedyk ER, Jones D, Critchley HO, Phipps RP, Blieden TM, Springer TA. Expression of stromal-derived factor-1 is decreased by IL-1 and TNF and in dermal wound healing. J Immunol. 2001;166:5749–54.
Jiang L, Zhu YQ, Du R, Gu YX, Xia L, Qin F, et al. The expression and role of stromal cell-derived factor-1alpha-CXCR4 axis in human dental pulp. J Endod. 2008;34:939–44.
de Brito LC, Teles FR, Teles RP, Totola AH, Vieira LQ, Sobrinho AP. T-lymphocyte and cytokine expression in human inflammatory periapical lesions. J Endod. 2012;38:481–5.
Kortesidis A, Zannettino A, Isenmann S, Shi S, Lapidot T, Gronthos S. Stromal-derived factor-1 promotes the growth, survival, and development of human bone marrow stromal stem cells. Blood. 2005;105:3793–801.
Jiang L, Peng WW, Li LF, Yang Y, Zhu YQ. Isolation and identification of CXCR4-positive cells from human dental pulp cells. J Endod. 2012;38:791–5.
Bartold PM, McCulloch CA, Narayanan AS, Pitaru S. Tissue engineering: a new paradigm for periodontal regeneration based on molecular and cell biology. Periodontol 2000. 2000;24:253–69.
Beertsen W, McCulloch CA, Sodek J. The periodontal ligament: a unique, multifunctional connective tissue. Periodontol 2000. 1997;13:20–40.
Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149–55.
Garcia-Ramallo E, Marques T, Prats N, Beleta J, Kunkel SL, Godessart N. Resident cell chemokine expression serves as the major mechanism for leukocyte recruitment during local inflammation. J Immunol. 2002;169:6467–73.
Acknowledgements
We would like to thank Dr. Toyama and Dr. Sato for P. gingivalis infection and the staff of the animal facility for care of the rats. We thank Prof. Hamada and Prof. Arakawa for fruitful discussions; Prof. Kook for P. gingivalis LPS; Dr. Bhawal for histological and microscopic analysis; Dr. Fujita for technical assistance.
Funding
Not applicable.
Availability of data and materials
The clinical data and personal details will not be made available in order to protect the participants’ identity.
Authors’ contributions
SJ performed the experiments and wrote the manuscript. HG analyzed the data. NE conceived and designed the experiments, critical review of manuscript. SK supervised interpretation of the data and critical review of manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was reviewed and approved by the Committee of Ethics on Human Experiments of the Dalian Stomatological Hospital (DLKQLL2015001; 11/18/2015). Written informed consent was obtained for all subjects after the nature and possible consequences of the studies were explained. The experimental procedures of this study were approved by the Committee of Ethics on Animal Experiments of the Kanagawa Dental University (97; 02/25/2010).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Sun, J., Nemoto, E., Hong, G. et al. Modulation of stromal cell-derived factor 1 alpha (SDF-1α) and its receptor CXCR4 in Porphyromonas gingivalis-induced periodontal inflammation. BMC Oral Health 17, 26 (2017). https://doi.org/10.1186/s12903-016-0250-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12903-016-0250-8