Abstract
Backgrounds
MIR31 host gene (MIR31HG) polymorphisms play important roles in the occurrence of osteonecrosis. However, the association of MIR31HG polymorphisms with the risk of steroid-induced osteonecrosis of the femoral head (SONFH) remains unclear. In this study, we aimed to investigate the correlation between MIR31HG polymorphisms and SONFH susceptibility in the Chinese Han population.
Methods
A total of 708 volunteers were recruited to detect the effect of seven single nucleotide polymorphisms (SNPs) in the MIR31HG gene on SONFH risk in the Chinese Han population. Genotyping of MIR31HG polymorphisms was performed using the Agena MassARRAY platform. The odds ratio (OR) and 95% confidence interval (95% CI) were used to evaluate the correlation between MIR31HG polymorphisms and SONFH risk using logistic regression model.
Results
According to the results of genetic model, rs10965059 in MIR31HG was significantly correlated with the susceptibility to SONFH (OR = 0.56, p = 0.002). Interestingly, the stratified analysis showed that rs10965059 was associated with the reduced risk of SONFH in subjects aged > 40 years (OR = 0.30, p < 0.001) and male populations (OR = 0.35, p < 0 .001). Moreover, rs10965059 was associated with the reduced risk of bilateral SONFH (OR = 0.50, p = 0.002). Finally, multi-factor dimension reduction (MDR) results showed that the combination of rs1332184, rs72703442, rs2025327, rs55683539, rs2181559, rs10965059 and rs10965064 was the best model for predicting SONFH occurrence (p < 0.0001).
Conclusion
The study indicated that rs10965059 could be involved in SONFH occurrence in the Chinese Han population, which might provide clues for investigating the role of MIR31HG in the pathogenesis of SONFH.
Similar content being viewed by others
Introduction
Osteonecrosis of the femoral head (ONFH) is a progressive rupture of the femoral head caused by the death of bone cells from various causes. The main characteristics of ONFH are the differentiation and the damage of bone marrow mesenchymal cells, enhanced cytotoxicity and destruction of vascular blood flow [1]. Osteonecrosis can be summarized into two categories: traumatic and non-traumatic femoral head necrosis. The steroid-induced osteonecrosis of the femoral head (SONFH), a non-traumatic femoral head necrosis, is a devastating disease, which is often result in devastating and crippling health conditions following steroid therapy [2]. In China, there were approximately 8 million patients with non-traumatic ONFH, which may be closely related to their frequent use of high-dose hormone therapy [3]. The pathogenesis is likely multifactorial, with genetic and environmental factors playing a role. Corticosteroid use, alcohol consumption, smoking, and infection and metabolic disease are all risk factors for SONFH [4]. Furthermore, genetics appears to play an important role in the development of SONFH. Previous studies have suggested that some genes play a role in SONFH occurrence including eNOS, PAI-1, VEGF, and ApoA [5]. However, there are still a large number of potential osteonecrosis-related genes and loci that have not been fully explored.
Long non-coding RNA (LncRNA) is a non-protein coding RNA molecule with a structure size of 200 nucleotides [6, 7]. Yuan and Sun’s studies showed that, lncRNA can regulate the development of immune diseases and affect immune function and autoimmunity, such as osteosarcoma and IgA nephropathy (IgAN) [8, 9]. The lncRNA MIR31 host gene (MIR31HG) is an crucial regulator of malignant tumors [10]. MIR31HG located on chromosome 9 with the length of 2166 bp, is an lncRNA that acts on the progression of cancers, such as osteosarcoma, lung cancer, breast cancer and cervical cancer [9, 11, 12]. For example, recent studies have mentioned that MIR31HG is also involved in the development and regeneration of bone, and the pathogenesis of numerous orthopaedic conditions [13]. MIR31HG was up-regulated in osteosarcoma (OS) tissues and OS cell lines. In the case of bone loss, it was usually inflamed in the defective or injured tissue. A previous research has demonstrated that knock-down of MIR31HG not only affects the enhancement of osteogenic differentiation, but also limits the inhibitory effect of osteogenic in an inflammatory environment [14]. Studies have found that the interference with MIR31HG can improve osteogenesis in bone marrow stromal cells in patients with cleidocranial dysplasia (BMSCs-CCD), possibly through by promoting osteogenic differentiation and improving the aging-related properties of BMSCs-CCD [15]. MIR31HG can regulate the tumor suppressor miR-361 and its target genes, and promote tumor progression in osteosarcoma acting as an oncogene [9]. These studies have shown that MIR31HG may play an important role in SONFH. At present, the connection between MIR31HG gene polymorphism and the susceptibility to SONFH was not reported.
In the case-control study, the MassARRAY platform was used to select seven single nucleotide polymorphisms (SNPs) in MIR31HG for genotyping. We further investigated the effect of MIR31HG genetic polymorphisms on SONFH risk and conducted the stratified analysis to identify the contribution of confounding factors to the association between SNPs and the risk of SONFH. Our research will provide a new perspective to study the role of MIR31HG on the susceptibility to SNOFH.
Methods
Subjects
A total of 708 unrelated participants were recruited, embracing 200 SONFH cases (41.15 ± 12.90 years) and 508 healthy controls (42.70 ± 13.01 years) geographically and ethnically matched. Exclusion criteria were as follows: (1) Patients who did not meet the diagnostic criteria of SONFH and patients with traumatic ONFH, hip dislocation and other hip diseases; and (2) Patients without major family genetic diseases. The histopathological diagnosis was based on X-rays and/or magnetic resonance imaging (MRI) examination of the hip and frog positions. The research protocol was in compliance with the Declaration of Helsinki and was approved by the ethics committee of Affiliated Hospital of Weifang Medical University and Second Affiliated Hospital of Inner Mongolia Medical University. All experimental subjects signed a written informed consent. Demographic and blood biochemical indicators of each subject were collected from standardized questionnaires and medical records by trained research staff.
Selection and genotyping for MIR31HG polymorphisms
Seven functional SNPs in MIR31HG (rs1332184, rs72703442, rs2025327, rs55683539, rs2181559, rs10965059 and rs10965064) were selected from the 1000 Genomes Project (http://www.1000genomes.org/), with the minor allele frequency (MAF) of each SNP greater than 0.05. Peripheral blood genomic DNA was extracted from all subjects according to the operating procedures of Whole Blood Genomic DNA Isolation Kit (Xi’an GoldMag Biotechnology, China). Agena MassARRAY iPLEX platform was used for genotyping. Agena Bioscience Assay was used to design PCR primers for amplification (Supplementary Table 1). Finally, Agena Bioscience TYPER application software 4.0 was performed to analyze the genetic data.
Statistical analysis
The differences in demographic or clinical characteristics between the case and the control groups were compared by χ2tests for the categorical variables and Student’s t-tests for continuous variables. PLINK software was used to detect four genetic models (co-dominant, dominant, recessive, and log-additive). Hardy-Weinberg equilibrium (HWE) of all SNPs from control individuals was evaluated by χ2 test. Multi-factor dimension reduction (MDR) is suitable for detecting the interaction between SNP-SNP and SONFH risk. Analysis of variance (ANOVA) was performed to determine differences in clinical characteristics among SNPs genotypes. By calculating the odds ratio (OR) and 95% confidence interval (CI), logistic regression results were adjusted for age and gender to assess the impact of MIR31HG polymorphism on SONFH risk. HaploView software version 4.2 and logistic regression were carried out to assess the correlation of MIR31HG haplotypes with SONFH susceptibility. All statistics were two-tailed, and a p < 0.05 was considered statistically significant. SPSS 20.0 software (Chicago, IL, USA) was used for statistical analysis in this study.
Results
Basic conventional characteristics
were consisted. The basic characteristics of 200 patients with SONFH and 508 healthy participants are summarized in Table 1, including age, gender, necrosis, and course. The mean age was 41.15 ± 12.90 years in the case group and 42.70 ± 13.01 years in control group. There were no significant differences in age (p = 0.152) and gender (p = 0.706) characteristics between cases and controls.
Association analysis of MIR31HG and SONFH risk
In this study, seven SNPs (rs1332184, rs72703442, rs2025327, rs55683539, rs2181559, rs10965059, and rs10965064) were successfully genotyped. The minor allele frequencies are record in Table 2. All SNP distribution of controls were in line with HWE (p > 0.05). The rs10965059-T allele frequency in the case group (0.103) was lower than that in the control group (0.169), and the reduced risk of SONFH was found (OR = 0.56, p = 0.002).
The correlation between the risk of SONFH and the MIR31HG polymorphisms was assessed after adjusting for age and gender in the four genetic models (co-dominant, dominant, recessive, and log-addition models). Our analysis results showed that rs10965059 was significantly related to the risk of SONFH (Table 3). In addition, rs10965059 was associated with the reduced susceptibility to SONFH in the co-dominant (T/C vs. C/C, OR = 0.50, p = 0.002), dominant (T/T-T/C vs. C/C, OR = 0.54, p = 0.004) and log-additive (OR = 0.63, p = 0.016) models.
Stratification analyses
To further investigate the effect of confounding factors on the association of MIR31HG variants with SONFH occurrence, we conducted a stratified analysis based on age, gender, disease course, and bilateral. The age-stratified analysis of the relationship between SNPs and SONFH risk is presented in Table 4. In subjects aged > 40 years, rs10965059 was related to a reduced risk of SONFH (T allele: OR = 0.30, p < 0.001; C/T genotype: OR = 0.34, p < 0.001; C/T-T/T genotype: OR = 0.33, p < 0.001). Conversely, no significant relationship of MIR31HG variants with SONFH risk was observed in subjects aged less than 40 years.
Gender-based stratified analysis (Table 5) indicated that, rs10965059 was a protective SNP for SONFH in males (T allele: OR = 0.53, p = 0.009; C/T genotype: OR = 0.35, p < 0.001; C/T-T/T genotype: OR = 0.42, p = 0.001).
Additionally, the stratification analysis of the association between MIR31HG SNPs and SONFH risk by course and bilateral are presented in Table 6. We discovered that rs2025327 might contribute to prolonged SONFH course (OR = 2.14, p = 0.046). The rs10965059 was associated with the reduced risk of bilateral SONFH (T allele, OR = 0.56, p = 0.005; C/T genotype, OR = 0.43, p = 0.002; C/T-T/T genotype, OR = 0.51, p = 0.007).
MDR analysis of SNP-SNP interaction on SONFH
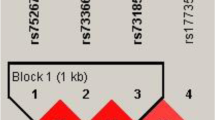
SNP-SNP interaction was determined using MDR analysis. As shown in Table 7 and Fig. 1, the analysis results indicated that the combination of rs1332184, rs72703442, rs2025327, rs55683539, rs2181559, rs10965059, and rs10965064 was the optimal model for predicting SONFH occurrence (training accuracy = 0.671, CVC = 10/10, p < 0.0001). In addition, the optimal single locus model for predicting SONFH risk was rs10965059 (training accuracy = 0.578, CVC = 10/10, p < 0.0001). Two-locus model was rs2181559 and 10,965,059. Three-locus model was consisted of rs2025327, rs2181559, and rs10965059. Four--locus model was consisted of rs1332184, rs2181559, rs10965059 and rs10965064. Five--locus model was the combination of rs1332184, rs72703442, rs55683539, rs2181559, rs10965059 and rs10965064. The results of the network diagram and the tree diagram were consistent (Fig. 1). There was a stronger redundant interaction between rs10965059 and rs10965064 (information gain: − 0.78%) and a stronger synergy between rs72703442 and rs2025327 (information gain: 0.26%).
The correlation of MIR31HG haplotypes with SONFH susceptibility
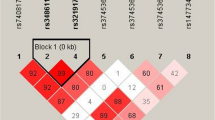
We also examined the impacts of MIR31HG haplotypes on SONFH susceptibility. As shown in Fig. 2, a linkage disequilibrium (LD) block was comprised of three SNPs including rs72703442, rs2025327 and rs55683539. The frequency distribution of haplotypes in case and control group is presented in Table 8. To examine the effect of haplotypes on SONFH risk, a haplotype-based logistic regression method was carried out in the case–control cohort, however, no significant association was found.
Discussion
SONFH is multi-layered and intricate disease with femoral neck fracture or bone tissue disorder, whose symptoms and signs are diverse, and the time and degree of pain attack are different However, SONFH still has the basis of pathological evolution. There are no specific clinical manifestations of ONFH, so it is difficult to make a diagnosis of ONFH from the patient’s symptoms and clinical examination [16]. With the development of modern precision medicine in recent years, an in-depth research on stem cells, molecular biology, and the exact pathogenesis of SONFH has been analyzed. A large number of experiments have shown that the increase in reactive oxygen species (ROS) caused by hormone use is related to the occurrence and development of SONFH [17, 18]. The the frequent collapse of the femoral head and hip joint dysfunction makes the treatment of SONFH difficult [4, 19].
LncRNA consists of a non-protein coding transcripts with approximately 200 nucleotides [6], which are drawn into various cellular processes such as chromatin remodeling, post-transcriptional processing and transcription process [20], involved in the occurrence, progression, and metastasis of human cancers, and played corresponding roles. Among those cancers, lncRNAs are more widely researched in osteosarcoma, including lncRNA-21A, UCA1, MEG3, HULC, and MIR31HG [21]. MIR31HG acts as an oncogene in osteosarcoma to promote tumor progression via regulation of tumor suppressor miR-361 and its target genes [9, 21]. Taken together, studying SONFH in the field of exploring lncRNAs is highly needed and promising. Moreover, according to our current research results, there is a significant correlation between MIR31HG polymorphism and SONFH susceptibility in the Chinese Han population.
MIR31HG is a kind of lncRNA that can be expressed in human bone cells, and it involves autoimmune in the recent research reports. The most noteworthy thing is that there is no research on the correlation between MIR31HG polymorphism and SONFH susceptibility. Our research is the first to find a significant risk connection between MIR31HG genetic variations and SONFH susceptibility in the Han population in China. The locus in MIR31HG has only been reported in IgAN currently [8]. In future studies, if SNP assessment is used as a type of risk marker, patients with high risk of ONFH can be identified through screening, and the dosage of steroids then can be differentiated based on individual differences, which can prevent the development of SONFH [1].
Therefore, we are committed to investigating the association between the MIR31HG gene polymorphism and the risk of SONFH disease. Our study results of genotyping showed that rs10965059-T allele frequency in the case group (0.103) was lower than that in the control group (0.169), and the reduced risk of SONFH was found. The stratified analysis results showed that rs10965059 was associated with the reduced risk of SONFH in subjects aged > 40 years (p < 0.001), and males (p < 0 .001). Consequently, we speculated that age and gender may interact with MIR31HG genetic polymorphisms on SONFH occurrence. Moreover, rs10965059 was associated with the reduced risk of bilateral SONFH (p = 0.002).
However, there are other candidate genes in the research on SONFH, and the research on MIR31HG is relatively rare. Nonetheless, our current work has some limitations. First of all, the relationship between SNPs and SONFH risk was investigated in the early stage, and the relationship among gene-environment interactions needs to be studied in the later work. Second, we have successfully demonstrated the relationship between MIR31HG polymorphisms and SONFH, and the molecular mechanism of SONFH will be studied in the future work. Patients were all from Shandong, Inner Mongolia and adjacent areas, which are in low population mobility. It is easy to carry out population-based research. As is known to all, this is the first study to probe into the effect of MIR31HG mutation on SONFH, which may provide a scientific basis for future research of MIR31HG on the molecular mechanism of SONFH.
Conclusion
Our study indicates that rs10965059 in MIR31HG is a protective SNP for SONFH, which offers a new insight for the molecular mechanism and provides a new major candidate gene in the study progression of SONFH. In the future, we will continue to collect samples to expand the sample size for confirming our results in a larger cohort of subjects.
Availability of data and materials
The datasets generated and/or analysed during the current study are not publicly available due [REASON WHY DATA ARE NOT PUBLIC] but are available from the corresponding author on reasonable request.
References
Wang Y, Li X, Gao Y, Li Z, Yu L, Meng Q, et al. Genetic polymorphisms of CYP3A4 among Chinese patients with steroid-induced osteonecrosis of the femoral head. Medicine. 2016;95(44):e5332.
Kubo T, Fujioka M, Ishida M. Clinical condition of steroid-induced osteonecrosis of the femoral head. Clin Calcium. 2007;17(6):856–62.
Zhao D-W, Yu M, Hu K, Wang W, Yang L, Wang B-J, et al. Prevalence of nontraumatic osteonecrosis of the femoral head and its associated risk factors in the Chinese population: results from a nationally representative survey. Chin Med J. 2015;128(21):2843–50.
Wang A, Ren M, Wang J. The pathogenesis of steroid-induced osteonecrosis of the femoral head: a systematic review of the literature. Gene. 2018;671:103–9.
Chang C, Greenspan A, Gershwin ME. The pathogenesis, diagnosis and clinical manifestations of steroid-induced osteonecrosis. J Autoimmun. 2020;110:102460.
Rigoutsos I, Lee SK, Nam SY, Anfossi S, Pasculli B, Pichler M, et al. N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration. Genome Biol. 2017;18(1):98.
Ling H, Vincent K, Pichler M, Fodde R, Berindan-Neagoe I, Slack FJ, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34(39):5003–11.
Yuan H, Li S, Wang L, Zhao X, Xue L, Lei X, et al. Genetic variants of the MIR31HG gene are related to a risk of IgA nephropathy. Int Immunopharmacol. 2020;84:106533.
Sun Y, Jia X, Wang M, Deng Y. Long noncoding RNA MIR31HG abrogates the availability of tumor suppressor microRNA-361 for the growth of osteosarcoma. Cancer Manag Res. 2019;11:8055–64.
Zhou Y, Fan Y, Zhou X, Mou A, He Y, Wang F, et al. Significance of lncRNA MIR31HG in predicting the prognosis for Chinese patients with cancer: a meta-analysis. Biomark Med. 2020;14(4):303–16.
Zheng Y, Liu L, Shukla GC. A comprehensive review of web-based non-coding RNA resources for cancer research. Cancer Lett. 2017;407:1–8.
Patel JS, Hu M, Sinha G, Walker ND, Sherman LS, Gallagher A, et al. Non-coding RNA as mediators in microenvironment-breast cancer cell communication. Cancer Lett. 2016;380(1):289–95.
Li1* Z: Emerging roles of long non-coding RNAs in osteonecrosis of the femoral head. Am J Transl Res 2020, 15;12(9):5984-5991.
Jin C, Jia L, Huang Y, Zheng Y, Du N, Liu Y, et al. Inhibition of lncRNA MIR31HG promotes osteogenic differentiation of human adipose-derived stem cells. Stem Cells. 2016;34(11):2707–20.
OBC DJA, SS A, MS C, WR S. Glucocorticoids act directly on osteoclasts to increase their life span and reduce bone density. Endocrinology. 2006;147(12):5592–9.
MM A, JL C, HD S. Nontraumatic osteonecrosis of the femoral head: ten years later. J bone and joint surg Amer. 2006;88(5):1117–32.
Kai C, Yuhao L, Jianbo H, Nathan P, Chao W, Jacob K, et al. Steroid-induced osteonecrosis of the femoral head reveals enhanced reactive oxygen species and hyperactive osteoclasts. Int J Biol Sci. 2020;16(11):1888–900.
Yuxian C, Chun Z, Hua Z, Rongkai Z, Zhiqiang Y, Bangrong X, et al. Comparative serum proteome expression of the steroid-induced femoral head osteonecrosis in adults. Experimental and therapeutic medicine. 2015;9(1):77–83.
Jieli Du1. A single-nucleotide polymorphism in MMP9 is associated with decreased risk of steroid-induced osteonecrosis of the femoral head. Oncotarget. 2016;7(42):68434–41.
Yan H, Qi G, Ping DX, Xiangting W. Mechanism of long noncoding RNAs as transcriptional regulators in cancer. RNA Biol. 2020;17(11):1680–92.
Smolle MA, Pichler M. The role of long non-coding RNAs in osteosarcoma. Noncoding RNA. 2018;4(1):7.
Acknowledgments
The authors thank all participants and volunteers in this study.
Funding
This work was supported by 2020 The Doctoral Research Fund of Affiliated Hospital of Weifang Medical University (2020BSQD11) and Shandong Medical and Health Science and Technology Development Plan Project (20210407547 and 202004070595).
Author information
Authors and Affiliations
Contributions
Yuan Wang: drafting and revising important content; Yexin Wang, Da Liang, and Hongtao Hu: performed the experiments; Guangwei Li, Xiaoguang Meng and Bing Zhu: analyzed the data; Wei Zhong: conceived and designed the experiments. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The protocol for this study was approved by the Ethics Committee of the Affiliated Hospital of Weifang Medical University and Second Affiliated Hospital of Inner Mongolia Medical University, and was in line with the Helsinki declaration. And the participant’s signature informed consent was received.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplemental Table 1.
Primers used for this study
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, Y., Wang, Y., Liang, D. et al. MIR31HG polymorphisms are related to steroid-induced osteonecrosis of femoral head among Chinese Han population. BMC Musculoskelet Disord 23, 836 (2022). https://doi.org/10.1186/s12891-022-05785-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12891-022-05785-w