Abstract
Background
The HIF-1α/Notch signaling pathway regulates cell proliferation, apoptosis, and metabolism in the intervertebral discs (IVDs) and is implicated in disc degeneration. The nucleus pulposus (NP) is an important structure adjacent to the IVDs. However, the role of the HIF-1α/Notch signaling pathway in NP cells obtained from patients with different Modic changes (MCs) remains unclear. The purpose of the present study was to investigate the role of HIF-1α and components of the Notch pathway in the NP obtained from patients with various MCs.
Methods
A total of 85 NP tissue samples were collected from patients undergoing diskectomy for the treatment of low back pain. The NP tissues were divided into four groups based on the adjacent endplate degeneration, namely, MC I, II, III, and negative MC groups. The expression of HIF-1α and Notch-related components was measured and compared.
Results
The expression of HIF-1α, Notch1, and Notch2 was gradually increased in the MC I and MC II groups compared with that in the negative MC group. HIF-1α and Notch-related components were rarely detected in the MC III group.
Conclusions
The expression of HIF-1α/Notch increased in the NP cells of patients with MC I and MC II. HIF-1α and Notch-related components are potential biomarkers and the HIF-1α/Notch signaling pathway may serve as a promising therapeutic target for disc degeneration in patients with MCs.
Similar content being viewed by others
Background
Modic changes (MCs) are changes in the vertebral body marrow and endplate lesions that appear as visual signals on magnetic resonance imaging (MRI). MCs were first described and classified into 3 general types by Modic et al. in 1988 [1, 2]. Different Modic types may represent different stages of the same pathological process. Modic type 1 changes (MC I) are associated with fissuring of the cartilaginous endplate corresponding to endplate edema. Modic type 2 changes (MC II) reflect fatty replacement of the adjacent marrow. Modic type 3 lesions (MC III) are observed in vertebral bodies with sclerotic changes.
Previous studies investigating MCs have focused mainly on the imaging changes and the association with low back pain (LBP) [3, 4]. Repetitive loading and periodic injury resulting in inflammation are two major pathological factors that contribute to MCs [5, 6], and result in endplate changes and intervertebral disc (IVD) degeneration. The potential association between IVD degeneration and MCs requires further investigation. Therefore, the aim of the present study was to clarify the correlation between IVD degeneration and MCs, specifically by focusing on the changes in hypoxic biomarkers.
Hypoxia-inducible factor (HIF) is a master transcription factor that mediates the activation of coordinated cellular responses in order to adapt to hypoxic environments. The HIF proteins are heterodimers that consist of a regulable α-subunit and a conserved β-subunit, and can be subdivided into three types: HIF-1, HIF-2, and HIF-3 [7, 8]. The hypoxia response elements (HREs) are the final point of integration of signaling pathways regulated by oxygen [8, 9]. Briefly, α-subunits dimerize with β-subunits to bind to HREs, which activate the transcription of effector genes under hypoxic conditions. The rapid degeneration of α-subunits inhibits the transcriptional activity of HIFs under normoxic conditions [8, 9]. In nucleus pulposus (NP) cells, HIF-1 plays an important role in the regulation of biological behaviors, such as energy metabolism, matrix metabolism, radical dismutation, cell proliferation and survival [7, 10]. Risbud et al. [11] revealed that HIF-1α was stably expressed in human, rat, and sheep NP cells under hypoxic conditions, suggesting that HIF-1α may serve as a potential target for the prevention and treatment of IVD degeneration. Therefore, in the present study, HIF-1α was used as an index of ischemia and anoxia in NP cells in the IVDs.
Notch is a hypoxia-sensitive receptor protein that regulates the proliferation of progenitor cells. There are four types of Notch receptors, namely, Notch1, Notch2, Notch3, and Notch4. Several studies have investigated the relationship between HIF-1 and Notch in physiological and pathological conditions [12,13,14]. Hypoxia activates the Notch signaling pathway to maintain IVD cell proliferation and accelerates catabolism. In the NP and annulus fibrosus (AF) in rat disc tissue, the hypoxia-induced increase in Notch mRNA expression can be blocked by a Notch signaling inhibitor. Furthermore, the expression of the Notch target gene HES1 was induced by hypoxia. Moreover, inhibition of Notch signaling inhibited the proliferation of disc cells. Analysis of human degenerated disc tissue revealed that the expression of Notch signaling proteins was increased. Additionally, the increased expression of inflammatory cytokines promoted Notch signaling in degenerated discs [15]. Therefore, the HIF-1α/Notch signaling pathway plays an important role in disc degeneration, and may serve as a potential therapeutic target for the restoration of cell numbers in degenerative disc disease.
The present study investigated the expression of HIF-1α and Notch in the bulging discs adjacent to end plates with MCs. Furthermore, we explored the relationship between MCs and disc degeneration using imaging, biochemical, and immunohistochemical methods to determine whether the expression of HIF-1α and Notch may aid the diagnosis and treatment of degenerative disc diseases.
Methods
Human tissue collection
The Ethics Committee of Huazhong University of Science and Technology approved this study and waived the requirement for informed consent. A total of 85 surgical NP tissue samples were collected from patients undergoing diskectomy for the treatment of LBP between January 2013 and January 2016. Each sample was obtained from the protrusive region of the IVD. The average LBP intensity was documented on a 0–10 numerical rating scale (NRS) as follows: 0 = no pain and 10 = worst possible pain (Table 1).
As shown in Fig. 1, patients with MC I, MC II, and MC III were included in the present study according to the inclusion criteria for MCs on MRI. The exclusion criteria were as follows: mixed MCs, ankylosing spondylitis, scoliosis, vertebral fractures, lumbar spine infection, spinal tumors, metastatic lesions, and other spine-related diseases; diabetes, hypertension, and other relevant medical history; history of spinal surgery, smoking, alcoholism, or drug use; psychological disorders, mental disorders, and other systemic disorders.
Isolation and culture of NP cells
NP cells from three patients in each group (L4/5; mean age, 55.16 years; 4 males and 8 females) were isolated as previously described [16]. After isolation, cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and penicillin/streptomycin. For hypoxic culture, NP cells were cultured in a tri-gas incubator (Huaxi Electronics Technetronic Co., Ltd.) containing 1% O2, 5% CO2, and 94% N2 for 8 h.
Protein isolation and Western blotting
NP tissues from three patients in each group were extracted and immediately placed on ice. The tissues were washed with pre-cooled PBS and homogenized using RIPA buffer (Aspen, China) supplemented with phenylmethanesulfonyl fluoride and protease and phosphatase inhibitors (Aspen, China). Tissue lysates were sonicated on ice, and protein concentrations were determined using a BCA protein assay kit (Sigma). Total protein was separated via SDS-PAGE on 10% gels and subsequently transferred onto PVDF membranes (EMD Millipore Corporation, USA) at 250 mA. After blocking with 5% non-fat dried milk in TBST at room temperature for 1 h, the membranes were incubated with anti-Notch1 (Abcam, ab44986), anti-NICD (Abcam, ab83232), anti-Notch2 (CST, 5732), anti-Notch3 (CST, 5276), anti-Notch4 (CST, 2423), anti-HIF-1α (Novus, NB100–105), anti-HES1 (Abcam, ab49170), and anti-GAPDH (ProteinTech, 60,004–1-Ig) primary antibodies overnight at 4 °C. After washing with TBST three times, the protein bands were incubated using HRP-conjugated secondary antibodies (ProteinTech) at room temperature for 1 h. The protein bands were detected using enhanced chemiluminescence detection reagents.
RNA extraction and real-time polymerase chain reaction (RT-PCR)
Total RNA was isolated from NP tissues frozen in liquid nitrogen using the RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. The quantity of the total RNA extracted was determined using a spectrophotometer. Total RNA was reversed transcribed into complementary DNA using the First Strand cDNA Synthesis Kit (TAKARA, Japan) according to the manufacturer’s protocol. The forward and reverse primers used for quantitative PCR are presented in Table 2. Quantitative PCR was performed in triplicate in a 96-well plate using the KAPA SYBR FAST qPCR Kit Master Mix. Target gene mRNA levels were quantified using the 2−ΔΔCt method and normalized to the internal reference gene GAPDH.
Immunohistochemical analysis
Paraffin-embedded NP tissue sections were deparaffinized in xylene and rehydrated through graded concentrations of ethanol. The sections were incubated with rabbit primary antibodies targeting HIF-1α and NICD overnight at 4 °C. Pre-immune rabbit nonspecific IgG antibody was used as a negative control. Immunopositive NP cells were counted in random fields of view and the results were expressed as a percentage of the total cells.
Statistical analysis
All experiments were performed in triplicate. Data were analyzed using GraphPad Prism statistical software (version 7.00, La Jolla, CA, USA). The unpaired t-test or analysis of variance was used to compare the different groups, as applicable. Correlations between the MCs and facet joint or disc degeneration were evaluated using Spearman’s correlation analysis. P < 0.05 was considered to indicate a statistically significant difference.
Results
Histological analysis
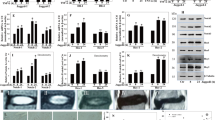
HIF-1α expression was analyzed in human NP tissues isolated from degenerated IVDs. The results revealed that NP tissues obtained from patients in the MC I and MC II groups expressed higher levels of HIF-1α protein than did those from the control group. The expression of HIF-1α protein in the MC III group was not significantly different from that in the control group (Fig. 2a). The percentage of cells positive for HIF-1α in the MC I and MC II groups was significantly higher than that in the control group (MC I: 33.96 vs. 9.8%, MC II: 41.52 vs. 9.8%, p < 0.01; Fig. 2b). The expression of Notch1 intracellular domain (NICD) was correlated to the increased expression of HIF-1α in the MC I and MC II groups (MC I: 29.75 vs. 18.32%, MC II: 41.96 vs. 18.32%, p < 0.01; Fig. 2c).
Gene expression
An increase in Notch1 and Notch2 mRNA levels was observed in the MC I and MC II groups, and this was positively correlated with upregulated HIF-1α expression. However, the expression of the aforementioned genes in patients with MC III was not statistically significantly different to that in the control group (Fig. 3a-c). Additionally, there was no significant difference in the expression of Notch3 in patients with MC I and MC III compared with that in the control group (Fig. 3d). The transcriptional level of Notch3 receptor in the MC II group was moderately increased compared with that in the control group. Levels of Notch4 transcripts in the MC groups were not significantly different to that in the control group (Fig. 3e). Moreover, the expression of HES1 (Fig. 3f), the target gene of Notch signaling, was significantly higher in the MC I and MC II groups than in the control group.
RT-PCR analysis of the mRNA levels of HIF-1α (a), Notch receptors (b-e), and HES1 (f) in tissues with different MCs. Spearman’s rank correlation analysis was performed to analyze the correlation between HIF-1α and Notch1 (g), HIF-1α and Notch2 (h), HIF-1α and Notch3 (i), and HIF-1α and Notch4 (j). Values are presented as the mean ± S.E.M. *p < 0.05, **p < 0.01
Correlation analysis
The expression of HIF-1α and Notch1 (Fig. 3g; p = 0.0001), HIF-1α and Notch2 (Fig. 3h; p = 0.0077), and HIF-1α and Notch3 (Fig. 3i; p = 0.0011), and HIF-1α and Notch4 (Fig. 3j; p = 0.0077) in patients with MC II were significantly correlated with one another (Table 3). No significant correlations were observed between the LBP NRS and HIF-1α or Notch receptor expression in any of the 4 groups (Table 4).
Protein expression in isolated NP cells
The isolated NP cells exhibited a HIF-1α-dependent increase in Notch1 and Notch2 protein levels (Fig. 4, p < 0.05). The expression levels of NICD and HES1 were significantly increased in the MC I and MC II groups compared with those in the control group, and this result was consistent with the trend observed for HIF-1α. These findings suggested that the expression of HIF-1α was correlated with the Notch signaling pathway in MC tissues.
Discussion
The present study revealed that the expression of HIF-1α was elevated in NP cells obtained from patients with MC I and MC II compared with that in cells obtained from patients with pure disc herniation. HIF-1α is an indicator of anaerobic states. In response to hypoxic conditions, cells increase the synthesis of HIF proteins [17]. HIF-1α plays an important role in the regulation of the biological behaviors of NP cells [7, 10]. Therefore, the increased expression of HIF-1α is indicative of degeneration, ischemia or hypoxia. Furthermore, previous studies revealed that the HIF-1α/Notch signaling pathway plays an important role in anoxic pathological processes that occur in tumors and neurodegenerative diseases [18, 19]. Based on the aforementioned studies, we hypothesized that there is a correlation between Notch and hypoxia, through HIF-1α expression, in patients with IVD herniation. Our study demonstrated that the expression of NICD in NP cells obtained from patients with IVD protrusion was increased, and was positively correlated with HIF-1α levels. Therefore, we speculated that HIF-1α may affect Notch signaling and downstream molecules in the NP cells of patients with disc herniation.
Based on the immunohistochemical results obtained in the present study, we assessed the mRNA expression levels of major components of the Notch signaling pathway in lumbar disc cells obtained from patients with different MC types. There was an increased expression of Notch1, NICD, and HES1 in the MC I and MC II groups, which was consistent with the histological results obtained. The correlation between HIF-1α and the Notch signaling pathway suggests that HIF-1α regulates disc regeneration through activation of the Notch-HES1 pathway. Specifically, HIF-1α may promote recruitment of the NICD to the CSL-binding motifs in the HES1 promoter and maintain homeostasis of the extracellular matrix in the NP [20, 21].
Spearman’s rank correlation analysis was performed to assess the correlation between the NRS score and gene expression levels of HIF-1α and Notch receptors. Unlike the imaging methods used to evaluate the degree of IVD degeneration [22], the biochemical markers investigated in the present study were not positively correlated with the clinical symptoms of LBP in the different MC groups. This inconsistent result is likely because the Notch-HES1 pathway is not involved in the initiation of LBP. A previous study revealed that there were significant correlations between LBP and inflammatory factors such as IL-6, IL-8, PGE2, and TNFα [23, 24]. The majority of these factors have been successfully used to activate Notch signaling in NP [15, 25]. In addition, HIF-1α expression was significantly increased in IL-1β-stimulated NP cells under hypoxic conditions [26]. Moreover, extensive research shows that LBP may be the result of a low-grade infection caused by Cutibacterium acnes in MC I [27, 28]. The hypoxic microenvironment of IVD provides a favorable condition for the growth of anaerobic bacteria, thereby facilitating consistent accumulation of inflammatory cytokines (IL-8, MIP-1α, MCP-1, IP-10, TNF-α) [29]. We therefore speculated that upregulated inflammatory factors and low-grade bacterial infection participate in the activation of the Notch and HIF-1α pathways and subsequent initiation of IVD degeneration in patients with MCs, particularly MC I and MC II [30].
Several studies have demonstrated crosstalk between the HIF-1α and Notch signaling pathways in IVD. To the best of our knowledge, the present study was the first to elucidate the co-expression patterns of the HIF-1α and the Notch signaling pathway in patients with different MCs. Specifically, hypoxia-induced Notch receptors and downstream molecules were highly expressed in patients with MC I and MC II, but not MC III, as detected by RT-PCR, western blotting, and immunohistochemistry. Furthermore, Notch1 and Notch2 mRNA levels were markedly elevated in the NP, while Notch3 and Notch4 levels were not altered as a result of the change in oxygen concentrations in IVDs with MCs.
Collectively, the results of the present study revealed that the HIF-1α and Notch signaling pathways play an important role in IVD degeneration. Therefore, these pathways may serve as novel therapeutic targets, particularly for patients who are ineligible for surgery. Prior to future clinical application, further investigation of the interaction between HIF-1α and Notch signaling and the influence of downstream molecules is required.
This study had certain limitations. Firstly, the small sample size in the MC I and MC III groups could have led to a large error when conducting the Spearman’s rank correlation analysis, and may have influenced the statistical correlation between HIF-1α and Notch1/Notch2 in the MC I and MC III groups. Secondly, as the AF and endplate (EP) sections of the IVD samples were too small to allow follow-up analysis, they were carefully excluded from the NP tissue. Therefore, we did not evaluate changes in gene and protein expression in AF and endplate tissues, and further studies are required. Furthermore, the samples used for western blotting must exhibit strong proliferative ability in vitro. The samples from L4/5 in each group may lead to a large margin of selection bias.
Availability of data and materials
Available upon request from the corresponding author.
Abbreviations
- IVD:
-
Intervertebral disc
- MCs:
-
Modic changes
- HIF-1α:
-
Hypoxia-inducible factor-1α
- NP:
-
Nucleus pulposus
- MRI:
-
Magnetic resonance images
- LBP:
-
Low back pain
- NICD:
-
Notch1 intracellular domain
- AF:
-
Annulus fibrosus
References
Modic MT, Steinberg PM, Ross JS, Masaryk TJ, Carter JR. Degenerative disk disease: assessment of changes in vertebral body marrow with MR imaging. Radiology. 1988;166(1 Pt 1):193–9.
Modic MT, Masaryk TJ, Ross JS, Carter JR. Imaging of degenerative disk disease. Radiology. 1988;168(1):177–86.
Vital JM, Gille O, Pointillart V, Pedram M, Bacon P, Razanabola F, Schaelderle C, Azzouz S. Course of Modic 1 six months after lumbar posterior osteosynthesis. Spine. 2003;28(7):715–20.
Schmid G, Witteler A, Willburger R, Kuhnen C, Jergas M, Koester O. Lumbar disk herniation: correlation of histologic findings with marrow signal intensity changes in vertebral endplates at MR imaging. Radiology. 2004;231(2):352–8.
Hansson T, Roos B. Microcalluses of the trabeculae in lumbar vertebrae and their relation to the bone mineral content. Spine. 1981;6(4):375–80.
Crock HV. Internal disc disruption. A challenge to disc prolapse fifty years on. Spine. 1986;11(6):650–3.
Boskey AL. Signaling in response to hypoxia and Normoxia in the intervertebral disc. Arthritis Rheum. 2008;58(12):3637–9.
Semenza GL. Signal transduction to hypoxia-inducible factor 1. Biochem Pharmacol. 2002;64(5–6):993–8.
Kaufman B, Scharf O, Arbeit J, Ashcroft M, Brown JM, Bruick RK, Chapman JD, Evans SM, Giaccia AJ, Harris AL, et al. Proceedings of the oxygen homeostasis/hypoxia meeting. Cancer Res. 2004;64(9):3350–6.
Risbud MV, Schipani E, Shapiro IM. Hypoxic regulation of nucleus Pulposus cell survival from niche to notch. Am J Pathol. 2010;176(4):1577–83.
Risbud MV, Guttapalli A, Stokes DG, Hawkins D, Danielson KG, Schaer TP, Albert TJ, Shapiro IM. Nucleus pulposus cells express HIF-1 alpha under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J Cell Biochem. 2006;98(1):152–9.
Hiyama A, Skubutyte R, Markova D, Anderson DG, Yadla S, Sakai D, Mochida J, Albert TJ, Shapiro IM, Risbud MV. Hypoxia activates the notch signaling pathway in cells of the intervertebral disc Implications in degenerative disc disease. Arthritis Rheum. 2011;63(5):1355–64.
Zou J, Li P, Lu F, Liu N, Dai JJ, Ye JJ, Qu X, Sun XL, Ma DX, Park J, et al. Notch1 is required for hypoxia-induced proliferation, invasion and chemoresistance of T-cell acute lymphoblastic leukemia cells. J Hematol Oncol. 2013;6:13.
Wang XM, Mao XO, Xie L, Greenberg DA, Jin KL. Involvement of Notch1 signaling in neurogenesis in the subventricular zone of normal and ischemic rat brain in vivo. J Cereb Blood Flow Metab. 2009;29(10):1644–54.
Wang H, Tian Y, Wang JR, Phillips KLE, Binch ALA, Dunn S, Cross A, Chiverton N, Zheng ZM, Shapiro IM, et al. Inflammatory cytokines induce NOTCH signaling in nucleus Pulposus cells IMPLICATIONS IN INTERVERTEBRAL DISC DEGENERATION. J Biol Chem. 2013;288(23):16761–74.
Choi H, Merceron C, Mangiavini L, Seifert EL, Schipani E, Shapiro IM, Risbud MV. Hypoxia promotes noncanonical autophagy in nucleus pulposus cells independent of MTOR and HIF1A signaling. Autophagy. 2016;12(9):1631–46.
Wenger RH, Gassmann M. Oxygen(es) and the hypoxia-inducible factor-1. Biol Chem. 1997;378(7):609–16.
Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci U S A. 2008;105(17):6392–7.
Wang RS, Zhang YW, Zhang X, Liu RZ, Zhang X, Hong SG, Xia K, Xia JH, Zhang ZH, Xu HX. Transcriptional regulation of APH-1A and increased gamma-secretase cleavage of APP and Notch by HIF-1 and hypoxia. Faseb J. 2006;20(8):1275.
Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, Ruas JL, Poellinger L, Lendahl U, Bondesson M. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9(5):617–28.
Liu Z, Li C, Meng X, Bai Y, Qi J, Wang J, Zhou Q, Zhang W, Zhang X. Hypoxia-inducible factor-lalpha mediates aggrecan and collagen pi expression via NOTCH1 signaling in nucleus pulposus cells during intervertebral disc degeneration. Biochem Biophys Res Commun. 2017;488(3):554–61.
Xiao L, Ni CL, Shi JD, Wang ZR, Wang SC, Zhang JW, Lu AQ. Analysis of correlation between vertebral endplate change and lumbar disc degeneration. Med Sci Monitor. 2017;23:4932–8.
Burke JG, Watson RWG, McCormack D, Dowling FE, Walsh MG, Fitzpatrick JM. Intervertebral discs which cause low back pain secrete high levels of proinflammatory mediators. J Bone Joint Surg-Br Vol. 2002;84B(2):196–201.
Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol. 2014;10(1):44–56.
Zheng YX, Liu CC, Ni L, Liu ZY, Mirando AJ, Lin J, Saijilafu, Chen D, Hilton MJ, Li B, et al. Cell type-specific effects of notch signaling activation on intervertebral discs: Implications for intervertebral disc degeneration. J Cell Physiol. 2018;233(7):5431–40.
Kwon WK, Moon HJ, Kwon TH, Park YK, Kim JH. The role of hypoxia in angiogenesis and extracellular matrix regulation of intervertebral disc cells during inflammatory reactions. Neurosurgery. 2017;81(5):867–75.
Capoor MN, Ruzicka F, Machackova T, Jancalek R, Smrcka M, Schmitz JE, Hermanova M, Sana J, Michu E, Baird JC et al: Prevalence of Propionibacterium acnes in Intervertebral Discs of Patients Undergoing Lumbar Microdiscectomy: A Prospective Cross-Sectional Study. PLoS One. 2016;11(8):e0161676.
Stirling A, Worthington T, Rafiq M, Lambert PA, Elliott TSJ. Association between sciatica and Propionibacterium acnes. Lancet. 2001;357(9273):2024–5.
Yuan Y, Chen Y, Zhou Z, Jiao Y, Li C, Zheng Y, Lin Y, Xiao J, Chen Z, Cao P. Association between chronic inflammation and latent infection of Propionibacterium acnes in non-pyogenic degenerated intervertebral discs: a pilot study. Eur Spine J. 2018;27(10):2506–17.
Ohtori S, Inoue G, Ito T, Koshi T, Ozawa T, Doya H, Saito T, Moriya H, Takahashi K. Tumor necrosis factor-immunoreactive cells and PGP 9.5-immunoreactive nerve fibers in vertebral endplates of patients with discogenic low back pain and Modic type 1 or type 2 changes on MRI. Spine. 2006;31(9):1026–31.
Acknowledgments
Not applicable.
Funding
National Natural Science Foundation of China (NSFC, No. 81301085, No. 81873999, No. 81672158), National Key R&D Program of China (2016YFC1100100) grant funds were received to support this work. The funders had role in editing and final approval of the manuscript.
Author information
Authors and Affiliations
Contributions
Design of the experiment: ZX, and JD; collection of human samples: ZX, JD, and JGZ; cell experiments: ZX, and SY; data assessment: ZX, and JD; statistical analysis: ZX; writing of the manuscript: ZX, and JD; editing and final approval of the manuscript: XG, and JZ. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The Ethics Committee of Huazhong University of Science and Technology approved this study and waived the requirement for informed consent. The manuscript submitted does not contain information about medical device(s)/drug(s).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Western blot anlysis of samples from different MCs patients were treated with CoCl2 (100 μM) for 24 h, or cultured in hypoxia condition for 4, 8, or 12 h.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xiong, Z., Ding, J., Zhou, J. et al. Correlation between the HIF-1α/Notch signaling pathway and Modic changes in nucleus pulposus cells isolated from patients with low back pain. BMC Musculoskelet Disord 21, 500 (2020). https://doi.org/10.1186/s12891-020-03505-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12891-020-03505-w