Abstract
Background
In this study, we aimed to evaluate the clinical utility of Metagenomic Next-Generation sequencing (mNGS) on bronchoalveolar lavage fluid (BALF) in diagnosis of Lower Respiratory Tract Infections (LRTIs).
Methods
In this study, we retrospectively analyzed 186 hospitalized patients who were suspected with LRTIs and performed mNGS (DNA) test of BALF simultaneously at The Fifth Affiliated Hospital of Sun Yat-Sen University from March 2023 to August 2023. Suspected LRTI was based on LRTI related clinical manifestations or imaging examination. Among them, 155 patients had undergone conventional culture and mNGS (DNA) testing simultaneously. Finally, 138 cases (89.03%,138/155) were diagnosed as LRTI and 17 cases (10.97%,17/155) were diagnosed as non-LRTI. Both detecting rate and diagnostic efficacy of mNGS and conventional culture were compared.
Results
The positive detection rates of pathogens between mNGS and conventional culture were significant different (81.29% VS 39.35%, P < 0.05). Compared with paired conventional culture result, the sensitivity of mNGS in diagnosis of LRTIs was more superior (88.41% VS 43.48%; P < 0.05), the specificity was opposite (76.47% VS 94.12%; P > 0.05). Furthermore, 77.54% and 35.51% of LRTI cases were being etiologically diagnosed by mNGS and culture respectively. Importantly, mNGS directly led to a change of treatment regimen in 58 (37.42%) cases, including antibiotic adjustment (29.68%) and ruling out active infection (7.74%). Moreover, treatment regimen remained unchanged in 97 (62.58%) cases, considering the current antibiotic therapy already covered the detected pathogens (36.13%) or empirical treatment was effective (11.61%).
Conclusions
mNGS can identify a wide range of pathogens in LRTIs, with improved sensitivity and being more superior at diagnosing LRTIs etiologically. mNGS has the potential to enhance clinical outcomes by optimizing the treatment regimens.
Similar content being viewed by others
Background
Lower Respiratory Tract Infections (LRTIs) continue to be a leading cause of death worldwide [1, 2]. Globally, a large-scale study estimated that 2.18 million deaths caused by LRTIs in 2021 [3, 4]. A variety of pathogens can be causes of LRTIs [3, 5], but the similar clinical manifestations among the patients infected by various pathogens keep the clinicians from making the accurate and timely etiological diagnosis, which will delay the administration of proper antibiotics, causing crucial effect on the outcomes of patients [6]. On the other hand, due to underlying diseases, compromised immune status, and abuse of wide-spectrum antibiotics in patients, especially elderly patients, the spectrum of the pathogens in the lower respiratory tract have been changed significantly and continuously which undoubtedly will be obstacles to seek for the causative pathogens.
Pathogen diagnosis for LRTIs has relied on conventional culture which limit to bacteria and fungi. However, it is a time-consuming procedure taking several days to complete. And the detection sensitivity can be seriously affected by the previous antibiotic use [7]. On the other hand, polymerase chain reaction (PCR) and immunology techniques are supplement tools for identification of microorganisms when culture fails. However, both of them are limited to specific pathogens, therefore the unexpected, rare or novel pathogens would be overlooked. In China, it is reported that only 30–60% of patients with LRTIs were diagnosed by conventional methods etiologically including culture, PCR testing and immunology testing [8]. Hence, new diagnostic methods for early and accurate etiological detection in LRTIs are in need urgently.
The emergence of Metagenomic Next-generation sequencing (mNGS) can be a promising technique to solve the problem. mNGS is an untargeted molecular method that can theoretically detect all pathogens unbiasedly in a single test and has revolutionized pathogen detection in clinical settings [9]. mNGS has the ability to detect a wide range of pathogens including bacteria, fungi, viruses, and parasites simultaneously and is less affected by prior antibiotic use [10]. There are many studies have explored the microbiological diagnostic performance of mNGS in LRTIs using BALF or sputum, especially for detection of atypical pathogens [11,12,13,14] and mix infections [15,16,17]. However, the high sensitivity of mNGS is accompanied by high false positive rate, which means not all detected pathogens are causative for the patient’s infection. Thus, accurate etiological diagnosis relies on the cautious interpretation of the mNGS result. However, there is no uniform standard to date for interpreting the mNGS result, and it is still a problem need to be solved [6, 17,18,19].
In this study, we retrospectively reviewed the mNGS results of the BALF samples from 186 adult patients with suspected LRTIs. The result of mNGS was compared with the paired culture performed concurrently. The diagnostic efficacy of BALF-mNGS was compared with the conventional culture. Additionally, the adjustment of clinical strategy based on the mNGS result were also analyzed to evaluate the clinical efficacy.
Methods
Patients and sample collection
We retrospectively summarized those patients suspected with Lower Respiratory Tract Infections (LRTIs) and performed BALF mNGS test simultaneously from March 2023 to August 2023 at The Fifth Affiliated Hospital of Sun Yat-Sen university. Suspected LRTIs were based on the clinical manifestations such as cough, chest pain, dyspnea, worsening of existing respiratory symptoms or infectious lesion indicated by imaging examination results judged by the physician. The inclusion criteria were as following: (1) Patients who were suspected with LRTIs; (2) Patients who performed the mNGS examination of BALF; (3) Patients with complete medical information. The exclusion criteria were as following: (1) Patients who refused to conduct the mNGS examination; (2) Aged under 18 years old; (3) Patients with incomplete medical information; (4) Samples from the same patients, the first sampling result was included, others were excluded. Finally, 186 patients were included in the study. The patients were from department of Respiratory(N = 53, 28.49%), Infectious disease(N = 51, 27.42%), ICU(N = 44, 23.66%), Thoracic surgery(N = 16, 8.60%), Hematology (N = 4, 2.15%), Rheumatology (N = 4, 2.15%), Transplantation (N = 4, 2.15%), Thoracic oncology (N = 3,1.61%), Geriatrics (N = 2, 1.08%), Neurology (N = 1, 0.54%), Breast disease (N = 1, 0.54%), Neurointerventional (N = 1, 0.54%), Cerebrovascular disease (N = 1, 0.54%), General medicine (N = 1, 0.54%), as shown in Table 1. All the outcomes were measured within a week after bronchoscopy.
Metagenomic next-generation sequencing and analysis
Sample processing and DNA extraction
1.5-3mL BALF was collected from each patient according to standard procedures. We extracted the DNA from 0.5 ml sample.
Construction of DNA libraries: DNA libraries were constructed through DNA fragmentation (about 200 bp), end repair, adapter ligation, PCR amplification, and DNB (DNA nanoballs) construction. DNB construction is conducted as following: the pooling DNA is denatured under 95℃ to form single-strand DNA, the single-strand DNA is cycled under 37℃. The cycle DNA is further amplified to make DNB. DNBs was qualified by Qubit ssDNA Assay Kit. Then, libraries with confirmed quality were sequenced by the MGISEQ-2000 platform (BGI Co.,Ltd, Shenzhen, China).
Sequencing and bioinformatic analysis
High-quality sequencing data were generated by removing low-quality reads, followed by computational subtraction of human host sequences mapped to the human reference genome (hg19). The remaining data by removal of low-complexity reads were classified by simultaneously aligning to PMDB Database, including 10,989 bacteria (196 Mycobacterium, Mycoplasma, Chlamydia and Rickettsia 159 in all), 1179 Fungi, 5050 viruses and 282 parasites.
Threshold criteria for interpretation of mNGS result
SMRN: number of sequences strictly mapped at species level. The raw result is mapped to the background microbiology database which consists of two parts: common contaminant microorganisms reported by clinical investigation and those microorganisms frequently detected in more than 50% samples in one month, the latter part is updated routinely. For different types of microorganisms, the thresholds were set as following: Bacterial/fungus/mycoplasma/chlamydia/Rickettsia/Virus: SMRN ≥ 3; Parasite: SMRN ≥ 100; Mycobacterium: SMRN ≥ 1. For microorganisms judged as candidate after the former step, SMRN of candidate microorganisms in sample is compared to the negative control group, only those higher than in the negative control group were retained. Furthermore, for SMRN ≤ 50, only those higher than 5 times in negative control group are reported, for SMRN > 50, only those higher than 3 times in negative control group are reported.
Defining the causative pathogens for LRTIs by mNGS
As reported in previous report [20]: we searched all micro-organisms detected by mNGS against PubMed to determine whether they caused pneumonia. Micro-organisms that could not cause pneumonia, including Corynebacterium, coagulase-negative Staphylococci, Neisseria, and anaerobic bacteria, which are considered normally parasitic in the human oropharynx in most cases in this study. After excluding the normal flora of the respiratory tract, pathogens detected by mNGS were categorized into four groups: (1) definite, BALF mNGS result is concordant with results from microbiologic tests (BALF culture, nucleic acid-based testing) performed within 7 days of BALF collection; (2) probable, BALF mNGS-based pathogen is likely the cause of Lower Respiratory Tract Infection based on clinical, radiologic, or laboratory findings; (3) possible, BALF mNGS-based pathogen has pathogenic potential and is consistent with clinical presentation but an alternate explanation is more likely; (4) unlikely, BALF mNGS-based pathogen has pathogenic potential but is not consistent with clinical presentation. As for culture, pathogens detected by culture were categorized into three groups: (1) definite, BALF culture-based pathogen is likely the cause of Lower Respiratory Tract Infection based on clinical, radiologic, or laboratory findings; (2) false positive: BALF culture-based pathogen has pathogenic potential but is not likely the cause of Lower Respiratory Tract Infection based on clinical, radiologic, or laboratory findings; (3) negative: BALF culture failed to detect any pathogen. The diagnosis of Lower Respiratory Tract Infection was made by doctor-in-charge based on a composite reference standard incorporating all testing results and clinical adjudication.
Statistical analysis
Categorical variables were compared using the chi-square test or Fisher’s exact test. P < 0.05 was considered significant. Continuous variables were presented as mean ± SEM. Statistical analyses were performed using GraphPad Prism 9 and SPSS statistics 26.
Results
Baseline features of patients
A total of 186 patients (107 male,57.53%;79 female,42.47%) with suspected Lower Respiratory Tract Infections (LRTIs) were included in this study (Table 1). The mean age was 58.59 ± 1.15 years old and the majority of patients were aged between 40-70 years old (61.29%). Among them, majority patients (73.12%) had underlying diseases, including 31(16.67%) cases with respiratory disease such as COPD and bronchiectasis, 36 (19.35%) cases with diabetes, 76 (40.86%) cases with cardiovascular disease such as hypertension, coronary heart disease and cerebral infarction, and 24(12.90%) cases with solid cancer. Moreover, in terms of immune status, 48 (25.81%) cases were immune-compromised, including condition with Rheumatic disease, Hematology disease, AIDS, Organ transplantation and Solid cancer. 159 cases (85.48%) were presented with LRTIs symptoms, while 27 cases (14.52%) were presented solely with abnormal image result without any symptom. 167 cases (89.78%) were reported with inflammation infiltration, while 19 cases (10.22%) were reported to be normal according to image examination result. 166 (89.25%) cases were diagnosed with LRTI. The baseline laboratory test including WBC, NEU, LYM, CRP and PCT were listed as follows, the average value of WBC, NEU, LYM are in the normal range while the average of CRP and PCT are above normal range. The patients were mainly from department of Respiratory, Infectious disease, ICU and Thoracic surgery. (Table 1)
Pathogen detection spectrum identified by mNGS
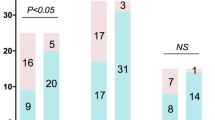
According to the mNGS result, 148 cases (148/186, 79.57%) were reported positive for at least one pathogen while 38 cases (38/186, 20.43%) were negative for all detected pathogens. Among the positive cases, 54 cases (54/148, 36.49%) were identified as single infection and 94 cases (94/148, 63.51%) were identified as mix infection categorized by pathogen species. Among the 155 cases with paired result of conventional culture, 24 and 28 cases were positive for bacteria and fungi respectively, 9 cases were detected with bacteria and fungi, while 94 cases were negative by culture (Fig. 1a). Among the mix infection detected by mNGS, “Bacteria + Virus”, “Bacteria + Fungi”, and “Bacteria + Fungi + Virus” accounted for 87.23% (82/94) of the mix infection cases. The frequently detected pathogens were also listed (Fig. 1b). Among positive result detected by mNGS, 113 cases (113/148, 76.35%) were positive for bacteria, 66 cases (66/148, 44.59%) were positive for fungi, 86 cases (86/148, 58.11%) were positive for virus. 6 cases of mycoplasma and 2 cases of chlamydia were identified by mNGS. For conventional culture, 33 cases and 37 cases were identified as positive for bacteria and fungi respectively (Fig. 1c). In a word, mix infection is more common than single infection and tend to be recognized by mNGS.
Infection type identified by mNGS and conventional culture. (a) Distribution of single and mix infection in 186 patients detected by mNGS and 155 patients with paired conventional culture result. (b) Comparison of pathogen detection of mix infection by mNGS. (c) Distribution of infection types sorting by species in 148 patients positive detected by mNGS and 61 patients positive detected by conventional culture
Among pathogens detected by mNGS, Pseudomonas aeruginosa (n = 22) was the most frequently detected Gram-negative bacteria, followed by Acinetobacter baumannii (n = 17), Klebsiella pneumoniae (n = 17), Stenotrophomonas maltophilia (n = 12) and Haemophilus influenzae (n = 10). As for Gram-positive bacteria, Mycobacterium tuberculosis was most frequently detected (n = 18), followed by Staphylococcus aureus (n = 16), Enterococcus faecium (n = 12), Enterococcus faecalis (n = 12), Corynebacterium striatum (n = 12). Moreover, 6 cases were detected with Nontuberculosis Mycobacteria (NTM), including Mycobacterium abscessus (n = 3), Mycobacterium Kansasii (n = 1), Mycobacterium colombiense (n = 1) and Mycobacterium Simiae (n = 1) (Fig. 2a). As for Fungi, Candida albicans (n = 28) ranked first, followed by Pneumocystis jirovecii (n = 15), Candida parapsilosis (n = 9), Aspergillus fumigatus (n = 7), and Candida glabrata (n = 6). As for virus, Human gammaherpesvirus 4 (EBV) was the most frequently detected virus (n = 42), followed by Torque teno virus (n = 29), Human betaherpesvirus 7 (n = 24), Human betaherpesvirus 5 (CMV) (n = 24), and Human alphaherpesvirus 1 (HSV-1) (n = 13). Additonally, 6 cases were detected with Mycoplasma including Mycoplasma hominis (n = 3), Mycoplasma pneumoniae (n = 2), Ureaplasma Urealyticum (n = 1). 2 cases were detected with Chlamydia including Mycoplasma Pneumoniae (n = 1) and Chlamydia psittaci (n = 1) (Fig. 2b). Generally, mNGS exhibits a much wider detection spectrum than conventional culture.
Comparison between mNGS and conventional culture testing
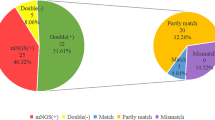
Among the 186 patients, both BALF-mNGS and conventional culture were conducted for 155 patients (155/186, 83.33%) while for another 31 patients (31/186, 16.7%), the BALF culture were not conducted or the result were incomplete. We focus on the 155 cases with both result of mNGS and conventional culture. When compared the results of these two detecting tools, 69 cases (69/155, 44.52%) were only positively detected by mNGS, 57 cases (57/155, 36.77%) were positive for both mNGS and conventional culture, 4 cases (4/155,2.58%) were only positively detected by conventional culture, and 25 cases (25/155, 16.13%) were negative for both tools (Fig. 3a). There was no wonder that the positive rate was higher in mNGS (81.29% VS 39.35%) as it detected more pathogens such as virus, Mycoplasma and Chlamydia. However, similar tendency (70.32% VS 39.35%) was also observed when limited to bacteria and fungi (Fig. 3b, middle panel), among the 53 cases positive for both mNGS and conventional culture, we compared the concordance of detected bacteria and fungi by these tools. The result of mNGS and conventional culture were perfect matched in 6 cases (6/53, 11.32%) and 8 cases (8/53, 15.09%) were totally unmatched. 39 cases (39/53, 73.58%) were found to be partial matched which means at least one detected pathogen was overlapped between the result of mNGS and conventional culture (Fig. 3b, right panel). On the other hand, 38 cases were detected negative for bacteria and fungi by both tools, 25 cases were negative for all detected pathogens, but 13 cases were detected with virus and atypical pathogens, including Mycoplasma and Chlamydia (Fig. 3b, left panel).
Comparison of pathogenic detection between mNGS and conventional culture. a. Pie chart showing the positive distribution of mNGS (including all detected pathogens by mNGS) and culture for 155 cases with paired testing result. b. Concordance between mNGS and conventional culture. Middle panel: Pie chart showing the positive distribution of mNGS (including bacteria and Fungi) and culture for 155 cases with paired testing result. Left panel: For the double-negative cases, 25(65.79%) cases were negative for all detected pathogens and 13 (34.21%) cases were detected with virus or atypical pathogens. Right panel: For the double-positive cases, majority cases (N = 39, 73.58%) were partial matching between the mNGS and culture result (at least one pathogen identified in the test was confirmed by the other), 6 cases (N = 6, 11.32%) were complete matched and 8 cases (N = 8, 15.09%) were unmatched
Comparison of clinical utility between mNGS and conventional culture
Among the 155 cases with result of both mNGS and conventional culture, 138 cases (138/155, 89.03%) were diagnosed as LRTI and 17 cases were not considered as LRTI (non-LRTI). The sensitivity and specificity of diagnosing Lower Respiratory Tract Infection by mNGS were 88.41% and 76.47%, respectively, with PPV and NPV being 96.83% and 44.83%. As for conventional culture, the sensitivity and specificity of diagnosing LRTI were 43.48% and 94.12%, respectively, with PPV and NPV being 98.36% and 17.02%. The combined test achieved a sensitivity of 90.58% and a specificity of 70.59%, with PPV and NPV being 96.15% and 48.00% (Table 2).
However, the detected pathogens are not equal to pathogens causing the infection, the discovery of the exact pathogens relies on interpreting the mNGS result combined with imaging examination, symptom and laboratory results. Among the 138 cases diagnosed as LRTI with results of mNGS and conventional culture, BALF mNGS detected 107 (77.54%) cases with define or probable pathogens which were likely the causative pathogens for the infection, while the causative pathogens of 49 (35.51%) cases were detected by conventional culture. However, mNGS failed to detected the specific causative pathogens in 16 (11.59%) cases with LRTI and 15 (10.87%) cases though being positive detected by mNGS, the detected pathogens were not considered the causative pathogens (Table 3).
Additionally, we further evaluate the treatment adjustment based on the decision made by the doctor in charge after interpreting the mNGS result. The treatment strategies were maintained in 97 (62.58%) cases. Among the 97 cases, the medical team decided to maintain the initial treatment because the symptoms were relieved after the empirical therapy though the specific pathogens was not defined in 18 cases. The antibiotic regimens were also continued in 56 cases as the current regimens already covered the causative pathogens detected by mNGS. On the other hand, the antibiotic use were altered in 58 (37.42%) cases, including antibiotic adjustment to target specific pathogens (N = 46, 29.68%) or rule out LRTIs (N = 12, 7.74%) (Table 4).
Discussion
In this study, we retrospectively analyzed 186 patients who were suspected with Lower Respiratory Tract Infections (LRTIs) and with available mNGS result of BALF. The spectrum of pathogens being recognized was much more diverse than conventional culture. Among the double positive cases detected by both tools, majority (73.58%) of them were partial matched, indicating the necessity to analyze all species detected by both tools to identify the causative pathogens.
In terms of diagnostic efficacy, the sensitivity of mNGS in diagnosis of LRTIs was more superior than that of conventional culture (88.41% VS 43.48%; P < 0.05), the specificity was opposite (76.47% VS 94.12%; P > 0.05), which were similar as reported previously [21]. Furthermore, among 138 patients diagnosed with LRTI, 107 (77.54%) cases and 49 (35.51%) cases were etiologically diagnosed by mNGS and culture respectively. However, there were 16 and 15 LRTI cases were negative or being difficult to identify the causative pathogens by mNGS. Importantly, mNGS directly led to alteration of treatment regimen in 58 (37.42%) cases but treatment regimens were not altered in 97 (62.58%) cases. Although the outcome in patients with treatment adjustment seems better than those without treatment adjustment, it should be noticed that the latter group consists of more patients with critical illness, the severe pneumonia accounts for 27.84% (27/97), the poor outcome were inevitable.
The present study has certain limitations. Firstly, RNA virus was not detected. Secondly, nearly all patients were given empirical antibiotic treatment before sampling, the positive detection of conventional culture might be affected. Thirdly, this retrospective study was limited by the nonrandom patient selection, not all patients suspected with LRTIs were performed with BALF mNGS test and the sample size was relatively small. The pathogen spectrum presented here might different from that of suspected LRTI population in real world.
To sum up, mNGS exhibited better capability on pathogen detection than conventional culture in clinical setting. Moreover, mNGS have shown great advantages in making etiology diagnosis and provide valuable information to guide antibiotic use.
Data availability
In accordance with the ethics committee of The Fifth Affiliated Hospital of Sun Yat-Sen university related to protecting individual privacy, metagenomic sequencing data that support the findings of this study have been deposited at CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) with accession number CNP0006076 (https://db.cngb.org/search/project/CNP0006076/) for controlled access. The controlled-access sequencing data are however available upon reasonable request from the corresponding Data Access Committee.
References
Torres A, Cilloniz C, Niederman M, Menéndez R, Chalmers J, van der Wunderink R. Poll T: Pneumonia. Nat Reviews Disease Primers. 2021;7(1):25.
File T, Ramirez J. Community-Acquired Pneumonia. N Engl J Med. 2023;389(7):632–41.
Bender RG, Sirota SB, Swetschinski LR, Dominguez R-MV, Novotney A, Wool EE, Ikuta KS, Vongpradith A, Rogowski ELB, Doxey M et al. Global, regional, and national incidence and mortality burden of non-COVID-19 lower respiratory infections and aetiologies, 1990–2021: a systematic analysis from the Global Burden of Disease Study 2021. The Lancet Infectious Diseases 2024.
GBD 2016 Lower Respiratory Infections Collaborators. Estimates of the Global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the global burden of Disease Study 2016. Lancet Infect Dis. 2018;18(11):1191–210.
Kradin R, Digumarthy S. The pathology of pulmonary bacterial infection. Semin Diagn Pathol. 2017;34(6):498–509.
Diao Z, Han D, Zhang R, Li J. Metagenomics next-generation sequencing tests take the stage in the diagnosis of lower respiratory tract infections. J Adv Res. 2022;38:201–12.
Li H, Gao H, Meng H, Wang Q, Li S, Chen H, Li Y, Wang H. Detection of pulmonary infectious pathogens from lung biopsy tissues by Metagenomic Next-Generation sequencing. Front Cell Infect Microbiol. 2018;8:205.
Zhu Y, Tang X, Lu Y, Zhang J, Qu J. Contemporary Situation of Community-acquired Pneumonia in China: a systematic review. J Translational Intern Med. 2018;6(1):26–31.
Chiu C, Miller S. Clinical metagenomics. Nat Rev Genet. 2019;20(6):341–55.
Miao Q, Ma Y, Wang Q, Pan J, Zhang Y, Jin W, Yao Y, Su Y, Huang Y, Wang M, et al. Microbiological Diagnostic performance of Metagenomic Next-generation sequencing when Applied to Clinical Practice. Clin Infect Diseases: Official Publication Infect Dis Soc Am. 2018;67:S231–40.
Jin W, Miao Q, Wang M, Zhang Y, Ma Y, Huang Y, Wu H, Lin Y, Hu B, Pan J. A rare case of adrenal gland abscess due to anaerobes detected by metagenomic next-generation sequencing. Annals Translational Med. 2020;8(5):247.
Ge M, Gan M, Yan K, Xiao F, Yang L, Wu B, Xiao M, Ba Y, Zhang R, Wang J, et al. Combining metagenomic sequencing with whole Exome sequencing to optimize clinical strategies in neonates with a suspected central nervous system infection. Front Cell Infect Microbiol. 2021;11:671109.
van Boheemen S, van Rijn A, Pappas N, Carbo E, Vorderman R, Sidorov I, van Hof T, Mei P, Claas H, Kroes E. Retrospective validation of a metagenomic sequencing protocol for combined detection of RNA and DNA viruses using respiratory samples from Pediatric patients. J Mol Diagnostics: JMD. 2020;22(2):196–207.
Babiker A, Bradley H, Stittleburg V, Ingersoll J, Key A, Kraft C, Waggoner J, Piantadosi A. Metagenomic sequencing to detect respiratory viruses in persons under investigation for COVID-19. J Clin Microbiol 2020, 59(1).
Zheng Y, Qiu X, Wang T, Zhang J. The Diagnostic Value of Metagenomic Next-Generation sequencing in Lower respiratory tract infection. Front Cell Infect Microbiol. 2021;11:694756.
Liang M, Fan Y, Zhang D, Yang L, Wang X, Wang S, Xu J, Zhang J. Metagenomic next-generation sequencing for accurate diagnosis and management of lower respiratory tract infections. Int J Infect Diseases: IJID : Official Publication Int Soc Infect Dis. 2022;122:921–9.
Gaston D, Miller H, Fissel J, Jacobs E, Gough E, Wu J, Klein E, Carroll K, Simner P. Evaluation of Metagenomic and targeted next-generation sequencing workflows for detection of respiratory pathogens from Bronchoalveolar Lavage Fluid Specimens. J Clin Microbiol. 2022;60(7):e0052622.
Han D, Li Z, Li R, Tan P, Zhang R, Li J. mNGS in clinical microbiology laboratories: on the road to maturity. Crit Rev Microbiol. 2019;45:668–85.
Chen H, Zhan M, Liu S, Balloux F, Wang H. Unraveling the potential of metagenomic next-generation sequencing in infectious disease diagnosis: challenges and prospects. Sci Bull 2024.
Chen H, Yin Y, Gao H, Guo Y, Dong Z, Wang X, Zhang Y, Yang S, Peng Q, Liu Y, et al. Clinical utility of In-house Metagenomic Next-generation sequencing for the diagnosis of Lower Respiratory Tract Infections and analysis of the host Immune Response. Clin Infect Diseases: Official Publication Infect Dis Soc Am. 2020;71:S416–26.
Xie G, Zhao B, Wang X, Bao L, Xu Y, Ren X, Ji J, He T, Zhao H. Exploring the clinical utility of Metagenomic Next-Generation sequencing in the diagnosis of pulmonary infection. Infect Dis Therapy. 2021;10(3):1419–35.
Acknowledgements
Not applicable.
Funding
This work was supported by GuangDong Basic and Applied Basic Research Foundation (No. 2019A1515110566).
Author information
Authors and Affiliations
Contributions
Yanfen Zheng co-ordinated the project, collected data, performed analyses and wrote the manuscript. Wei Liu revised the manuscript. Tongyang Xiao collected data. Yan Liu and Hongtao Chen designed, supervised the project and reviewed the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The Study was conducted in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This Study was approved by the ethics committee of The Fifth Affiliated Hospital of Sun Yat-Sen university. The data were anonymous and informed consent was performed. Our study is registered in Chinese Clinical Trial Registry. The clinical trial number of this study is ChiCTR2400086468.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zheng, Y., Liu, W., Xiao, T. et al. Clinical utility of metagenomic next-generation sequencing on bronchoalveolar lavage fluid in diagnosis of lower respiratory tract infections. BMC Pulm Med 24, 422 (2024). https://doi.org/10.1186/s12890-024-03237-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12890-024-03237-w