Abstract
Background
Yanghe Pingchuan decoction (YPD) has been used for asthma treatment for many years in China. We sought to understand the mechanism of YPD, and find more potential targets for YPD-based treatment of asthma.
Methods
An ovalbumin-induced asthma model in rats was created. Staining (hematoxylin and eosin, Masson) was used to evaluate the treatment effect of YPD. RNA-sequencing was carried out to analyze global gene expression, and differentially expressed genes (DEGs) were identified. Analysis of the functional enrichment of genes was done using the Gene Ontology database (GO). Analysis of signaling-pathway enrichment of genes was done using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Real-time reverse transcription-quantitative polymerase chain reaction was undertaken to measure expression of DEGs.
Results
Pathology showed that YPD had an improvement effect on rats with asthma. RNA-sequencing showed that YPD led to upregulated and downregulated expression of many genes. The YPD-based control of asthma pathogenesis may be related to calcium ion (Ca2+) binding, inorganic cation transmembrane transporter activity, microtubule motor activity, and control of canonical signaling (e.g., peroxisome proliferator-activated receptor, calcium, cyclic adenosine monophosphate). Enrichment analyses suggested that asthma pathogenesis may be related to Ca2 + binding and contraction of vascular smooth muscle. A validation experiment showed that YPD could reduce the Ca2 + concentration by inhibiting the Angiopoietin-II (Ang-II)/Phospholipase (PLA)/calmodulin (CaM0 signaling axis.
Conclusion
Control of asthma pathogenesis by YPD may be related to inhibition of the Ang-II/PLA/CaM signaling axis, reduction of the Ca2+ concentration, and relaxation of airway smooth muscle (ASM).
Similar content being viewed by others
Introduction
Bronchial asthma is also referred to as ‘asthma’. Asthma is a chronic inflammatory disease of the airways caused by the participation of various inflammatory cells, inflammatory mediators, and neurotransmitters [1]. Asthma is a common respiratory disease with a high incidence. According to a report from the Global Initiative for Asthma Committee, among more than 300 million patients suffering from asthma worldwide, more than 30 million patients reside in China [2]. In China, the incidence of asthma in young children is high [3]. Asthma is one of the most common chronic diseases in the world, and its incidence is increasing worldwide, bringing a high economic burden to society [4].
The etiology of asthma is incompletely understood. Clinical studies have shown genetic factors and the environment to be the main risk factors for inducing asthma [5, 6]. The pathogenesis of asthma is relatively complex and not fully elucidated. Research on the pathogenesis of asthma has focused mainly on chronic inflammation of the airways, airway hyperresponsiveness [7], airway remodeling [8], and genetic mechanisms [9].
The main focus of treatment for asthma is symptomatic relief and control of further development of the disease [10]. The main drugs used for asthma treatment are glucocorticoids [11], leukotriene-receptor antagonists [12], and β2-receptor agonists [13]. However, long-term use of these agents may elicit adverse reactions [14]. For example, prolonged use of high-dose glucocorticoids increases the risk of bacterial infections [15], eye, pharyngeal, and other diseases in children, as can affect the growth and development of children.
Traditional Chinese medicine (TCM) has unique characteristics in the treatment of asthma. The therapy of asthma based on TCM concepts centers on relieving symptoms during an attack and treating its cause after remission. Yanghe Pingchuan decoction (YPD) is a TCM formulation and has been used to treat asthma. YPD is composed of Ephedra sinica Stapf (mahuang), Rehmannia glutinosa (Gaertn.) DC (shudihuang), Inula japonica Thunb (xuanfuhua), Morinda officinalis F.C.How (bajitian), Schisandra chinensis (Turcz.) Baill (wuweizi), Sinapis alba L (baijiezi), Draba nemorosa L (tinglizi), Angelica sinensis (Oliv.) Diels (danggui), Platycodon grandiflorum (Jacq.) A.DC (jiegeng). YPD have also been used in the clinical treatment of asthma for many years with clear curative effects [16]. Previously, we demonstrated that YPD has a therapeutic effect on rats with asthma, and that its mechanism of action may be related to inhibition of the phosphoinositide 3-kinase/protein kinase B signaling pathway [17]. However, the specific molecular mechanisms underlying the effects of YPD are incompletely understood.
Herein, we attempted to elucidate the potential molecular mechanisms underlying the effects of YPD on a rat model of asthma using RNA-sequencing.
Materials and methods
Chemical reagent
Ovalbumin (Lot: CLCB9757) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Calcium ion test kits were purchased from Jiancheng Bioengineering Research Institute (Nanjing, China). The RNA extraction kit were provided by Sangong Bioengineering (Shanghai Co., Ltd. China). Reverse Transcription Kit(with dsDNase) were provided by Biosharp (Hefei, China).
YPD preparation
All of herbs of YPD were obtained from Anqing Huashi Chinese Herbal Medicine Beverage (Anqing City, China). YPD preparation involved three main steps. First, each herb was immersed in water (10×) for 1 h, and decocted by boiling for 2 h. Second, after filtration, the residue was decocted with water (8×) for 1.5 h. Third, the two decoctions were combined and concentrated to 0.2 g/mL in a vacuum at 50 °C to obtain a YPD suspension.
Animal experiments and sample collection
The protocol for all animal experiments was approved (AHUCM-rats-2,022,093) by the Animal Experiment Ethics Committee of Anhui University of Chinese Medicine (Anhui, China).
All experimental methods and drug regimens were based on our previous work [17]. Male Sprague–Dawley rats (200 ± 20 g) were obtained from the Experimental Animal Center of Anhui Medical University. Rats were divided randomly into three groups of 10: control, asthma, and YPD. On day 1, rats in the asthma group and YPD group were injected (i.p.) with 10% ovalbumin and 1 mL of physiologic (0.9%) saline. On day 15, rats were placed in a box for nebulization (1% ovalbumin plus 0.9% saline, 400 mmHg, 15 min), once every 2 days for 10 days to create an asthma model. From day 28 of modeling, the YPD group was given YPD (14.76 g/kg, i.g.) and the control group and asthma group were given an identical amount of 0.9% saline, once a day, for 14 consecutive days. After the experiment, each rat was fasted for 8 h, and anesthetized with sodium pentobarbital [18, 19]. Blood was collected from the abdominal aorta, and samples were taken for testing.
Histopathology
After killing, right lungs were harvested, fixed, embedded in paraffin, and sectioned. Changes in the structure of airway walls and pathologic alterations in bronchi and smooth muscles were observed by staining (hematoxylin an eosin) and analyzed using Image J (US Institutes of Health, Bethesda, MD, USA).
Staining
Tissues were fixed in 4% paraformaldehyde at 55℃ for 1 h. The extent of bronchial fibrosis was determined by staining with Masson’s trichrome (1%) for 5–10 min at 55℃. Then, we evaluated the extent of fibrosis in lungs and bronchial tissue with an optical microscope and undertook pathology grading.
RNA extraction
Total RNA was extracted and purified by the mirVana™ miRNA Isolation Kit (catalog number: AM1561; Ambion, Austin, TX, USA) according to manufacturer instructions. RNA quality was evaluated by a spectrophotometer (Nanodrop™; Thermo Fisher Scientific, Waltham, MA, USA) and a bioanalyzer (2100 series; Agilent Technologies, Santa Clara, CA, USA).
Library preparation for transcriptome sequencing (RNA-sequencing)
RNA libraries were prepared using the SureSelect Chain-specific RNA Library Preparation kit (Agilent Technologies). Thereafter, a fluorometer (Qubit 3.0; Thermo Fisher Scientific) and bioanalyzer (2100 series; Agilent Technologies) were used to quantify the purified libraries. Cluster generation was undertaken using cBot (Illumina, San Diego, CA, USA). Sequencing was achieved with NovaSeq™ 6000 (Illumina). These procedures were outsourced to Hefei Yuan En Biotechnology Co., LTD (Hefei, China).
Differentially expressed genes (DEGs)
FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) [15] was used for the quality control of experimental samples to ensure that reads of low quality, high N content of unknown bases, or adapter contamination were filtered out. Hisat (http://daehwankimlab.github.io/hisat2/) [15] was employed to count the reads numbers mapped to genes. To make the expression of different genes and different samples comparable, based on the length of the gene itself and the amount of sequencing data, we converted the reads into fragments per kilobase of exon model per million mapped reads (FPKM). EdgeR (https://bioconductor.org/) was used to analyze DEGs between samples. After obtaining the p-value, correction of the multiple hypothesis test was undertaken. The threshold of the p-value was determined by controlling the false discovery rate (FDR), and the corrected p-value was the Q value. Simultaneously, we calculated the fold change (FC) based on the FPKM value. The conditions for DEG screening were Q ≤ 0.05 and FC ≥ 2.
Enrichment analyses
To understand more deeply the functions of DEGs, R (R Institute for Statistical Computing, Vienna, Austria) was used to analyze the enrichment of function based on the Gene Ontology (GO) database (http://geneontology.org/) [15] and enrichment of signaling pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (www.genome.jp/) [15]. We used clusterProfiler (https://bioconductor.org/) to analyze enrichment of function and signaling pathways. The top-30 enriched functions and signaling pathways were obtained. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/) [15] was employed to analyze protein–protein interaction (PPI) networks.
Validation experiment
Real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was used to verify the reliability of RNA-sequencing results. Transcriptome analysis showed the genes to have upregulated expression were Ang-II, PLA and CaM, so they were selected for validation by real-time RT-qPCR. β-actin was an internal reference gene. The 2−ΔΔ CT method was used to calculate the relative expression of genes.
RT-qPCR was applied to measure mRNA expression of Ang-II, PLA and CaM. Samples of total RNA were taken from right-lung tissues, seeded in a six-well culture plate at 1 × 106 cells/well, allowed to incubate for 24 h, and exposed to lysis buffer (Science Biotech, Beijing, China). Then, the absorbance at 260 and 280 nm of total RNA was determined by an ultraviolet spectrophotometer to determine the mRNA concentration. mRNA was reverse-transcribed into complementary-DNA with a cDNA reverse transcription kit according to manufacturer instructions. RT-qPCR was done on the LightCycler™ 480 II system (Roche Diagnostics, Basel, Germany/) using 2 × S6 Universal SYBR qPCR Mix (EnzyArtisan, Shanghai, China). The thermal cycling conditions were pre-denaturation at 95 °C for 30 s, denaturation at 95 °C for 10 s, annealing and extension at 60 °C for 30 s for 45 cycles, and extension at 72 °C for 5 min. The genes targeted for amplification were Ang-II, PLA and CaM, with β-actin as the internal reference gene. The 2−ΔΔCt method was applied to normalize the relative expression of mRNA. The primer sequences were devised and synthesized by EnzyArtisan, and are shown in Table 1.
Ca2+ was determined by methyl thymol blue (MTB) microplate method
We weighed tissue accurately. Nine times the volume of deionized water was added to tissue. The tissue homogenate was centrifuged (2500 × g, 10 min, room temperature). Then, 10% of the supernatant of the homogenate was removed for measurement. After cell samples had been collected, deionized water (0.3–0.5 mL) was added to 1 × 106 cells. After ultrasonic crushing, samples were mixed, centrifuged (2500 rpm, 10 min, room temperature), and the supernatant removed for measurement. A 96-well plate that had been treated with dilute hydrochloric acid and cleaned with deionized water was selected. Blank holes, standard holes, and measurement holes were assigned (Table 2) and solutions added in sequence.
Statistical analyses
Results were analyzed by SPSS 23.0 (IBM, Armonk, NY, USA). Data are the mean ± SD. One-way ANOVA was used to test for significant between-group differences. Differences were considered significant at p < 0.05 or p < 0.01. Differential analyses of GO and KEGG pathway enrichment were analyzed and results were considered statistically significant at p < 0.05. The conditions for DEG screening were p ≤ 0.05 and|log FC| ≥ 2.
Results
Therapeutic effect of YPD on rats suffering from asthma
In the control group, there was no infiltration of inflammatory cells to the bronchus, the bronchial wall was smooth, cells were arranged in a regular manner, and the thickness of the soft muscle layer was normal. Compared with the control group, there were many eosinophils, monocytes, and lymphocytes around the bronchial wall in the asthma group. Proliferation of airway epithelial cells, thickening of the smooth-muscle layer, as well as thickening and prolongation of the bronchial mucosa were also documented (Fig. 1a). After YPD treatment, the infiltration of inflammatory cells (eosinophils and lymphocytes) in rats with asthma was reduced significantly, and the thickness of the bronchial wall and smooth-muscle layer was close to that of the control group.
Staining (Masson’s trichrome) showed the structure of lung tissue in the control group to be normal with few collagen fibers (blue) or muscle fibers (red) [20] (Fig. 1b). Compared with the control group, the bronchial wall and alveolar wall were obviously thickened and consolidated in the model group, with a significant increase in the areas stained blue and red. After YPD treatment, the number of collagen fibers and muscle fibers was reduced significantly.
Identification of DEGs
RNA-sequencing was used to understand multifaceted mechanisms in rats with asthma [26]. We measured mRNA expression of each sample in control and asthma groups. The results of principal component analysis showed considerable separation between these two groups, indicating that the transcriptome sequencing was of high quality and could be analyzed further (Fig. 2A). A total of 812 DEGs were identified in the model group compared with the control group (Fig. 2B) (635 genes with upregulated expression and 177 genes with downregulated expression).
Analysis of DEGs with altered function after asthma onset
Classification of gene function can describe the properties of genes and products comprehensively, and such classification is updated constantly. Classification is based mainly on biological process (BP), cellular component (CC), and molecular function (MF). Comparison of data from analyses of functional enrichment with the genomic background aids understanding of which biological functions are associated significantly with DEGs. DEGs had to meet the criteria of corrected p-value (Q-value) ≤ 0.05. The biological functions of DEGs that met these criteria were determined.
We performed functional-enrichment analysis on DEGs with upregulated expression and downregulated expression in the asthma group: 635 genes had upregulated expression and 6230 GO terms were identified (4849 for BP, 528 for CC, and 853 for MF). The top-five GO terms with the smallest p-value (i.e., the most significant enrichment) were selected for each GO classification.
The top-five enriched functions for BP were ‘cilium organization’ (GO: 0044782), ‘cilium assembly’ (GO: 0060271), ‘cilium movement’ (GO: 0003341), ‘axoneme assembly’ (GO: 0035082), and ‘microtubule-based movement’ (GO: 0007018) (Fig. 3). The top-five enriched functions for CC were ‘cilium’ (GO: 0005929), ‘axoneme’ (GO: 0005930), ‘ciliary plasm‘ (GO: 0097014), ‘motile cilium’ (GO: 0031514), and ‘plasma membrane bounded cell projection cytoplasm’ (GO: 0032838). The top-five enriched functions for MF were ‘microtubule motor activity’ (GO: 0003777), ‘dynein intermediate chain binding’ (GO: 0045505), ‘ATP-dependent microtubule motor activity, minus-end-directed’ (GO: 0008569), ‘tubulin binding’ (GO: 0015631), and ‘dynein heavy chain binding’ (GO: 0045504).
The 177 genes with downregulated expression were enriched with 3788 GO terms (2919 for BP, 346 for CC, and 523 for MF). The top-five enriched functions for BP were ‘muscle system process’ (GO: 0003012), ‘muscle contraction’ (GO: 0006936), ‘striated muscle contraction’ (GO: 0006941), ‘striated muscle cell development’ (GO: 0055002), and ‘muscle cell development’ (GO: 0055001). The top-five enriched functions for CC were ‘contractile fiber’ (GO: 0043292), ‘myofibril’ (GO: 0030016), ‘sarcomere’ (GO: 0030017), ‘I band’ (GO: 0031674), and ‘actin cytoskeleton’ (GO: 0015629). The top-five enriched functions for MF were ’actin binding’ (GO: 0003779), ‘tropomyosin binding’ (GO: 0005523), ‘actin filament binding’ (GO: 0051015), ‘telethonin binding’ (GO: 0031433), and ‘metal ion transmembrane transporter activity’ (GO: 0046873).
Analysis of signaling pathways of DEGS altered after asthma onset
Analysis of signaling-pathway enrichment was done using the KEGG database. We discovered that 635 upregulated genes were enriched in 226 signaling pathways. The 10 most significantly enriched signaling pathways were ‘PPAR signaling pathway’ (FDR: 0.002001573), ‘regulation of lipolysis in adipocytes’ (FDR: 0.069244813), ‘AMPK signaling pathway’ (FDR: 0.069244813), ‘histidine metabolism’ (FDR: 0.069244813), ‘insulin signaling pathway’ (FDR: 0.069244813), ‘neuroactive ligand–receptor interaction’ (FDR: 0.074370072), ‘aldosterone synthesis and secretion’ (FDR: 0.120696471), ‘hepatitis C’ (FDR: 0.120696471), ‘tyrosine metabolism’ (FDR: 0.139915558), and ‘Huntington disease’ (FDR: 0.139915558). The smaller the FDR value, the more significant was the enrichment. We selected the top-20 signaling pathways with the smallest FDR value (Fig. 4A). The 177 genes with downregulated expression were enriched in 151 signaling pathway. The 10 most significantly enriched signaling pathways were ‘arrhythmogenic right ventricular cardiomyopathy’ (FDR: 0.057189133), ‘hypertrophic cardiomyopathy’ (FDR: 0.063430675), ‘cardiac muscle contraction’ (FDR: 0.21866133), ‘GABAergic synapse’ (FDR: 0.21866133), ‘dilated cardiomyopathy’ (FDR: 0.21866133), ‘serotonergic synapse’ (FDR: 0.334112021), ‘GnRH secretion’ (FDR: 0.334112021), ‘renin secretion’ (FDR: 0.395775367), ‘oxytocin signaling pathway’ (FDR: 0.525036286), and ‘adrenergic signaling in cardiomyocytes’ (FDR: 0.525036286). Based on the FDR value, we selected the top-20 most significantly enriched signaling pathways (Fig. 4B).
PPI network
Genes with significant differential expression were used to construct a PPI network based on the STRING database. The PPI network had 154 nodes and 189 edges (Fig. 5), among which the higher ranked degree values were recombinant myosin light chain 1 (MYL1), radical S-adenosyl methionine domain-containing protein 2 (RSAD2), interferon regulatory factor 7 (IRF7), and peroxisome proliferative activated receptor G (PPARG).
The top-five DEGs with upregulated expression and downregulated expression are listed in Table 3. We used the MCODE plugin in Cytoscape (https://cytoscape.org/) [27] to analyze the 12 modules obtained. Modules with a score > 5 and > 5 nodes were filtered and visualized according to ranking of the module score (Fig. 6). Most of DEGs belonging to the three modules had high expression in the asthma group.
RT-qPCR verification of DEGs
Differential expression of genes was detected by RT-qPCR (Fig. 7), Expression of Ang-II, PLA and CaM in the asthma group was increased significantly compared with that in the control group (p < 0.05). After intervention with YPD, expression of Ang-II, PLA and CaM in the YPD group was downregulated significantly compared with that in the asthma group (p < 0.05). These results suggested that YPD may inhibit the Ang-II/PLA/CaM signaling axis.
Effect of YPD on the intracellular concentration of Ca2+
A calcium detection kit based on use of methylthymol blue in microplates was used to measure the intracellular concentration of Ca2+. Compared with the control group, the Ca2+ concentration in the asthma group was increased significantly (p < 0.01) (Fig. 8). After YPD intervention, the Ca2+ concentration in the YPD group was downregulated significantly compared with that in the asthma group. These results suggested that YPD may reduce the intracellular Ca2+ concentration in rats suffering from asthma.
Discussion
YPD have been shown to be effective in controlling asthma, including inhibiting excessive proliferation of bronchial cells and alleviating inflammation [17, 18]. However, due to the multiple-component nature of TCM formulations, how YPD controls asthma in rats is not known.
We explored how YPD protects against asthma using RNA-sequencing. RNA-sequencing in the asthma group and control group revealed 812 DEGs, of which 635 genes had upregulated expression and 177 genes had downregulated expression, compared with the control group. Analyses of functional enrichment (using the GO database), signaling-pathway enrichment (using the KEGG database), and PPI networks provided deeper understanding of the pathologic mechanisms of asthma. We hypothesized that YPD may control asthma pathogenesis by binding Ca2+, inorganic cation transmembrane transporter activity, and microtubule motor activity. We also observed some canonical signaling pathways (e.g., peroxisome proliferator-activated receptor, calcium, cyclic adenosine monophosphate).
Asthma is induced by inflammation, remodeling, and hyperresponsiveness of the airways [21, 22]. Airway hyperreactivity includes intense constriction of the airways and airway spasm. Airway smooth muscle (ASM) cells are the main cell type that cause bronchial hyperresponsiveness [23], so they are also considered to be a target to combat bronchiectasis. In normal airways, ASM cells contract to regulate the airway diameter and bronchial motor tension. In asthma, there is a complex relationship between ASM cells and inflammatory cells (e.g., mast cells, T lymphocytes). Airway contraction is caused mainly by ASM contraction. There are many reasons for ASM contraction, but recent studies have shown that ASM contraction may be related to an increased level of Ca2+ [24, 25].
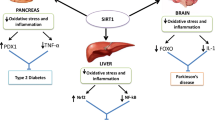
The current strategy to control an acute attack of asthma is to relax ASM and dilate the bronchus [26]. Ca2+ regulation is crucial to the contraction and relaxation of ASM [27]. If the Ca2+ concentration in ASM cells increases, Ca2+ binds to calmodulin and activates myosin light chain kinase (MLCK), resulting in MLC phosphorylation [28]. Phosphorylation of MLC increases the activity of myosin adenosine triphosphate (ATP)ase, which hydrolyzes ATP to release energy [29]. Actin-myosin is induced to transbridge circulation, resulting in shortened ASM. Therefore, regulation of the Ca2+ concentration in ASM cells is essential for controlling asthma symptoms. The intracellular Ca2+ concentration is regulated by calcium channels [30], ryanodine receptors [31], calcium pumps [32], and Na+/Ca2+ transporters [33], which can mediate the transmembrane flow of Ca2+. Ca2+-channel transport is one of the main ways in which Ca2+ enters cells. Ca2 channels in cell membranes can be divided into five categories (voltage-operated, voltage-dependent, receptor-operated, second messenger-operated, and mechanically operated). Among them, voltage-dependent Ca2 channels are the most important because they are ubiquitous in various tissues in the body, and are the main pathway of Ca2+ influx [34]. If a cell is depolarized, voltage-operated Ca2+ channels transfer Ca2+ from outside to inside the cell. Studies have shown that 20-hydroxyeicosatetraenoic acid (20-HETE) can cause the depolarization of vascular smooth muscle cells [35]. 20-HETE is an important metabolite in the cytochrome P450 metabolic pathway of arachidonic acid. The latter is stored in the cell membrane and released into the cytoplasm through regulation of phospholipase A [36]. Angiotensin can increase the PLA level through a series of reactions. Therefore, the angiotensin-2 signaling axis may have an important regulatory role in ASM contraction (Fig. 9, by Figdraw).
Analysis of signaling-pathway enrichment revealed five DEGs to be enriched with regard to contraction of vascular smooth muscle, among which expression of Ang-II, PLA and CaM was increased in the asthma group, and YPD could reverse this increase. Functional-enrichment analysis showed that YPD-based control of asthma pathogenesis may be related to Ca2+ binding and inorganic cation transmembrane transporter activity. Therefore, we measured expression of the Ang-II/PLA/CaM signaling axis in asthma and YPD treatment to ascertain if ASM is related to Ca2+ (the Ca2+ level in the all three groups was measured simultaneously). The Ang-II/PLA/CaM signaling axis was activated in the asthma group and the Ca2+ level increased; YPD could inhibit the Ang-II/PLA/CaM signaling axis and reduce the Ca2+ concentration.
Conclusions
We used deep RNA-sequencing to reveal that YPD could reduce the Ca2+ concentration by the Ang-II/PLA/CaM signaling axis in vitro. Our study provides insights into the potential applicability of YPD in asthma therapy.
Data availability
The datasets used and/or analysed during the current study areavailable from the GEO database ((http://www.ncbi.nlm.nih.gov/geo) [Accession number: GSE247032 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE247032)]. The original data are available from the corresponding author upon reasonable request.
Abbreviations
- YPD:
-
Yanghe Pingchuan decoction
- DEGs:
-
differentially expressed genes
- TCM:
-
Traditional Chinese medicine
- FDR:
-
false discovery rate
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- PPI:
-
protein–protein interaction
- RT-qPCR:
-
Real-time reverse transcription-quantitative polymerase chain reaction
- ASM:
-
Airway smooth muscle
References
Gkavogiannakis NA, Tsoporis JN, Drosatos IA, Tsirebolos G, Izhar S, Sakadakis E, Triantafyllis AS, Parker TG, Kalogiros LA, Leong-Poi H et al. Emergent inflammatory markers and echocardiographic indices in patients with bronchial asthma. Biomolecules 2023, 13(6).
Radhapyari L, Verma PK, Kumar V, Kumar M, Gupta AK, Bhat NK. Association between neutrophil lymphocyte ratio and status of symptom control in children and adolescents with bronchial asthma. Trop Doct 2023:494755231175709.
Wang J, Yang L, Sun P, Guo C, Jin Y, Jing X. Expression patterns of serum miR-27a-3p and activating transcription factor 3 in children with bronchial asthma and their correlations with airway inflammation. Clin Respir J 2023.
Freitas MM, Cavalcante PM, Duarte-Filho L, Macedo CAF, Brito MC, Menezes PMN, Ribeiro TF, Costa SM, Carvalho BAG, Ribeiro F, et al. Investigation of the relaxing effect of a camphor nanoemulsion on rat isolated trachea. Chemico-Biol Interact. 2021;348:109656.
Nair AK, Van Hulle CA, Bendlin BB, Zetterberg H, Blennow K, Wild N, Kollmorgen G, Suridjan I, Busse WW, Dean DC. Impact of asthma on the brain: evidence from diffusion MRI, CSF biomarkers and cognitive decline. Brain Commun. 2023;5(3):fcad180. 3rd et al.
Huang W, Schinasi LH, Kenyon CC, Auchincloss AH, Moore K, Melly S, Robinson LF, Forrest CB, De Roos AJ. Evaluation of evidence for interaction between PM2.5 and aeroallergens on childhood asthma exacerbation in Philadelphia, PA, 2011 to 2016. Environ Res 2023:116395.
Sun J, Bai S, Zhao J, Li D, Ma X, Ma L, Su X. Mapping knowledge structure and research of the biologic treatment of asthma: a bibliometric study. Front Immunol. 2023;14:1034755.
Ali NH, Rehman S, Naqvi M, Gulati K, Ray A. Modulation of immunological, biochemical, and histopathological changes of Airway Remodeling by Withania somnifera in an experimental model of allergic asthma in rats. J Pharmacopunct. 2023;26(2):158–66.
Kim HK, Kang JO, Lim JE, Ha TW, Jung HU, Lee WJ, Kim DJ, Baek EJ, Adcock IM, Chung KF, et al. Genetic differences according to onset age and lung function in asthma: a cluster analysis. Clin Translational Allergy. 2023;13(7):e12282.
Puxkandl V, Currie A, Hoetzenecker W, Altrichter S. Therapy resistant urticaria as a long-term symptom of an incomplete Schnitzler syndrome. Allergy Asthma Clin Immunology: Official J Can Soc Allergy Clin Immunol. 2023;19(1):64.
Domínguez-Ortega J, Delgado Romero J, Muñoz Gall X, Marco A, Blanco-Aparicio M. [Use of systemic glucocorticoids for the treatment of severe asthma: Spanish multidisciplinary Consensus]. Open Respiratory Archives. 2022;4(4):100202.
Vadagam N, Haridasyam SB, Venkatanarayana M, Lakka NS, Chinnakadoori SR. Separation and quantitative estimation of stereo-selective enantiomers of montelukast in pharmaceutical drug substance and tablets dosage forms by using stability-indicating normal phase-HPLC method. Chirality 2023.
Carvajal Gonczi CM, Hajiaghayi M, Gholizadeh F, Xavier Soares MA, Touma F, Lopez Naranjo C, Rios AJ, Pozzebon C, Daigneault T, Burchell-Reyes K et al. The β2-adrenergic receptor agonist terbutaline upregulates T helper-17 cells in a protein kinase A-dependent manner. Human immunology 2023.
He X, Chang Z, Yan H, Weng Y. Pulmonary aspergillus infection with abnormal imaging successfully treated with omalizumab: a case report. Medicine. 2023;102(24):e33845.
Ota R, Hata T, Hirata A, Hamada T, Nishihara M, Neo M, Katsumata T. Risk of infection from glucocorticoid and methotrexate interaction in patients with rheumatoid arthritis using biologics: a retrospective cohort study. Br J Clin Pharmacol. 2023;89(7):2168–78.
Yang L, Zhu H. Clinical study on Yang-He-Ping-Chuan granules treated chronic lung function duration particles bronchial asthma airway inflammation. J Chengdu Univ Traditional Chin Med. 2014;37(03):52–5.
Pan LY, Han YQ, Wang YZ, Chen QQ, Wu Y, Sun Y. Mechanism of Yanghe Pingchuan granules treatment for airway remodeling in asthma. Drug Des Devel Ther. 2018;12:1941–51.
Pan L, Chen Y, Jiang Y, Sun Y, Han Y, Wang Y. Yanghe Pingchuan Granules Alleviate Airway Inflammation in Bronchial Asthma and Inhibit Pyroptosis by Blocking the TLR4/NF-κB/NRLP3 Signaling Pathway. Mediators of inflammation 2022, 2022:6561048.
Gong C, Pan L, Jiang Y, Sun Y, Han Y, Wang D, Wang Y. Investigating the mechanism of action of Yanghe Pingchuan Granule in the treatment of bronchial asthma based on bioinformatics and experimental validation. Heliyon. 2023;9(11):e21936.
Balestrini A, Joseph V, Dourado M, Reese RM, Shields SD, Rouge L, Bravo DD, Chernov-Rogan T, Austin CD, Chen H et al. A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment. J Exp Med 2021, 218(4).
Simunovic M, Boyle J, Erbas B, Baker P, Davies JM. Airborne grass pollen and thunderstorms influence emergency department asthma presentations in a subtropical climate. Environ Res 2023:116754.
Dunbar H, Hawthorne IJ, Tunstead C, Armstrong ME, Donnelly SC, English K. Blockade of MIF biological activity ameliorates house dust mite-induced allergic airway inflammation in humanized MIF mice. FASEB Journal: Official Publication Federation Am Soc Experimental Biology. 2023;37(8):e23072.
Alqahtani T, Parveen S, Alghazwani Y, Alharbi HM, Gahtani RM, Hussain N, Rehman KU, Hussain M. Pharmacological validation for the Folklore Use of Ipomoea nil against Asthma: in vivo and in Vitro evaluation. Molecules 2022, 27(14).
Zeng Z, Cheng M, Li M, Wang T, Wen F, Sanderson MJ, Sneyd J, Shen Y, Chen J. Inherent differences of small airway contraction and ca(2+) oscillations in airway smooth muscle cells between BALB/c and C57BL/6 mouse strains. Front cell Dev Biology. 2023;11:1202573.
Huang AS, Tong BC, Hung HC, Wu AJ, Ho OK, Kong AH, Leung MM, Bai J, Fu X, Yu Z, et al. Targeting calcium signaling by inositol trisphosphate receptors: a novel mechanism for the anti-asthmatic effects of Houttuynia cordata. Biomed Pharmacotherapy = Biomedecine Pharmacotherapie. 2023;164:114935.
Satori NA, Pacini ESA, Godinho RO. Impact of the cAMP efflux and extracellular cAMP-adenosine pathway on airway smooth muscle relaxation induced by formoterol and phosphodiesterase inhibitors. Chemico-Biol Interact. 2023;382:110630.
Romero-Martínez BS, Sommer B, Solís-Chagoyán H, Calixto E, Aquino-Gálvez A, Jaimez R, Gomez-Verjan JC, González-Avila G, Flores-Soto E, Montaño LM. Estrogenic modulation of ionic channels, pumps and exchangers in Airway smooth muscle. Int J Mol Sci 2023, 24(9).
MacEwen MJS, Rusnac DV, Ermias H, Locke TM, Gizinski HE, Dexter JP, Sancak Y. Mathematical modeling and biochemical analysis support partially ordered calmodulin-myosin light chain kinase binding. iScience. 2023;26(4):106146.
Pires I, Hung YF, Bergmann U, Molloy JE, Kursula I. Analysis of Plasmodium Falciparum myosin B ATPase activity and structure in complex with the calmodulin-like domain of its light chain MLC-B. J Biol Chem. 2022;298(12):102634.
Rocha DG, Holanda TM, Braz HLB, de Moraes JAS, Marinho AD, Maia PHF, de Moraes MEA, de Fechine-Jamacaru FV. Moraes Filho MO: Vasorelaxant effect of Alpinia zerumbet’s essential oil on rat resistance artery involves blocking of calcium mobilization. Fitoterapia 2023:105623.
Caramia M, Romanov RA, Syderomenos S, Hevesi Z, Zhao M, Krasniakova M, Xu ZD, Harkany T, Hökfelt TGM. Neuronal diversity of neuropeptide signaling, including galanin, in the mouse locus coeruleus. Proc Natl Acad Sci USA. 2023;120(31):e2222095120.
Moysan L, Fazekas F, Fekete A, Köles L, Zelles T, Berekméri E. Ca(2+) Dynamics of Gap Junction Coupled and Uncoupled Deiters’ Cells in the Organ of Corti in Hearing BALB/c Mice. Int J Mol Sci 2023, 24(13).
Cheng PC, Cheng RC, Huang RC. Glutamate-evoked ca(2+) responses in the rat suprachiasmatic nucleus: involvement of na(+)/K(+)-ATPase and na(+)/Ca(2+)-Exchanger. Int J Mol Sci 2023, 24(7).
Ballarini E, Malacrida A, Rodriguez-Menendez V, Pozzi E, Canta A, Chiorazzi A, Monza L, Semperboni S, Meregalli C, Carozzi VA et al. Sodium-calcium exchanger 2: a pivotal role in Oxaliplatin Induced Peripheral neurotoxicity and axonal damage? Int J Mol Sci 2022, 23(17).
Agostinucci K, Hutcheson R, Hossain S, Pascale JV, Villegas E, Zhang F, Adebesin AM, Falck JR, Gupte S, Garcia V, et al. Blockade of 20-hydroxyeicosatetraenoic acid receptor lowers blood pressure and alters vascular function in mice with smooth muscle-specific overexpression of CYP4A12-20-HETE synthase. J Hypertens. 2022;40(3):498–511.
Barnabei L, Laplantine E, Mbongo W, Rieux-Laucat F, Weil R. NF-kappaB: at the Borders of autoimmunity and inflammation. Front Immunol. 2021;12:716469.
Acknowledgements
Not applicable.
Funding
This research was supported by the Anhui Provincial College Natural Science Research Key Project (No.2023AH050863, No.KJ2019A0440, No.KJ2020A0409).
Author information
Authors and Affiliations
Contributions
JJC and YZW conceived and designed the study. CXG and YHS performed the in vitro experiments. LYP and BFH wrote the manuscript, XCD and YQH provided ideas for the experimental design and modified the manuscripts to ensure the integrity of the entire experimental design. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The protocol for all animal experiments was approved (AHUCM-rats-2022093) by the Animal Experiment Ethics Committee of Anhui University of Chinese Medicine (Anhui, China). All the operations followed the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and ARRIVE Guidelines pertaining to animal experimentation.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Pan, L., He, B., Gong, C. et al. Analysis of global gene expression using RNA-sequencing reveals novel mechanism of Yanghe Pingchuan decoction in the treatment of asthma. BMC Pulm Med 24, 137 (2024). https://doi.org/10.1186/s12890-024-02952-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12890-024-02952-8