Abstract
Background
Hypoxic pulmonary hypertension (HPH) is a syndrome of abnormally elevated pulmonary artery pressure, and it is mostly caused by vasoconstriction and remodeling of the pulmonary artery induced by long-term chronic hypoxia. There is a high incidence of HPH, a short survival time of the patients, but currently no effective treatments.
Methods
In this study, HPH-related single cell sequencing (scRNA-seq) and bulk RNA sequencing (RNA-seq) data were downloaded from the public database of Gene Expression Omnibus (GEO) for bioinformatics analysis in order to find out genes with important regulatory roles in the development of HPH. 523 key genes were identified through cell subpopulation identification and trajectory analysis of the downloaded scRNA-seq data, and 41 key genes were identified through weighted correlation network analysis (WGCNA) of the bulk RNA-seq data. Three key genes: Hpgd, Npr3 and Fbln2 were identified by taking intersection of the key genes obtained above, and Hpgd was finally selected for subsequent verification. The human pulmonary artery endothelial cells (hPAECs) were treated with hypoxia for different periods of time, and it was found that the expression of Hpgd decreased in hypoxia-treated hPAECs in a time-dependent manner. In order to further confirm whether Hpgd affects the occurrence and development of HPH, Hpgd was overexpressed in hPAECs.
Results
Hpgd was confirmed to regulate the proliferation activity, apoptosis level, adhesiveness and angiogenesis ability of hypoxia-treated hPAECs through multiple experiments.
Conclusions
Downregulation of Hpgd can improve the proliferation activity, reduce apoptosis, and enhance adhesion and angiogenesis in endothelial cells (ECs), thus promoting the occurrence and development of HPH.
Similar content being viewed by others
Background
Pulmonary arterial hypertension (PAH) is a progressive occlusive pulmonary vascular disease caused by proliferation of endothelial cells (ECs) and smooth muscle cells (SMCs) as well as the occlusion of small peripheral pulmonary arteries induced by in situ thrombosis. There are multiple causes and a high mortality of PAH [1]. According to the classification of the World Health Organization (WHO), PAH can be divided into 5 subtypes, and hypoxic pulmonary hypertension (HPH), as one of the most common types, belongs to the third subtype [2]. HPH is commonly seen in chronic hypoxic lung diseases, including chronic obstructive pulmonary disease (COPD), obstructive sleep apnea syndrome, interstitial lung disease and chronic mountain sickness [3]. It is caused by hypoxia-induced endothelium injury, imbalance of vascular endothelial synthesis and secretion of various vasodilators, leading to early pulmonary vasoconstriction and pulmonary vascular remodeling. Therefore, it is believed that hypoxic pulmonary vascular remodeling is an important pathological basis of HPH [4]. The development of HPH is accompanied by inflammatory responses and oxidative stress [5], and is related to the worsening of symptoms and poor prognosis caused by elevated arterial pressure and right ventricular hypertrophy [6], but the exact pathogenesis of HPH is still unclear. At present, it has been clinically shown that long-term oxygen therapy can effectively prolong the survival of patients with COPD, but it can only slightly reduce the pulmonary artery pressure of patients [7]. Targeted drugs such as pulmonary vasodilators have not been shown to be effective in the clinical treatment of HPH [8]. Therefore, it is necessary to further explore the exact pathogenesis of HPH to provide more possibilities for the treatment of HPH.
ECs play important roles in the pathogenesis of HPH. Previous studies have demonstrated that up-regulation of HIF-1 and VEGF expression in ECs can induce vascular remodeling in HPH [9]. ECs and SMCs mediate vascular remodeling and PAH through FoxM1 signaling interactions [10]. The up-regulation of endothelial DKK1 (Dickkopf 1) can promote the development of PAH through the Sp1/SHMT2 pathway [11]. The heterogeneity of vascular ECs [12] may be a key factor for the ineffective treatment of HPH with current drugs [13], but fortunately, single cell sequencing (scRNA-seq) can well solve the problem of cell heterogeneity, so as to reveal the complex and rare cell population, the regulatory relationship between genes, and track the trajectories of different cell lineages during development. At present, a large number of studies were found to combinedly make use of scRNA-seq and bulk RNA-seq data for bioinformatics analysis [14, 15], and the results obtained complement each other, making the research more detailed and in-depth.
In this study, the scRNA-seq and bulk RNA-seq data were downloaded from the public database of Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) for joint analysis. Cell subpopulation identification and trajectory analysis were carried out on the scRNA-seq data, and differentially expressed genes (DEGs) analysis and WGCNA were conducted on the bulk RNA-seq data to find out the DEGs highly correlated with HPH. Three key genes: Hpgd, Npr3 and Fbln2 were obtained by taking intersection of the important genes obtained from the two datasets, and Hpgd was selected for subsequent verification. Hpgd was verified by Western blot and qRT-PCR, and was confirmed to be significantly down-regulated in hypoxia-treated human pulmonary artery endothelial cells (hPAECs). To further figure out the role of Hpgd in the pathogenesis of HPH, we overexpressed Hpgd in ECs in vitro, and measured ECs’ proliferative activity, apoptosis level, adhesion and angiogenesis. In a word, through bioinformatics analysis and experimental validation, this study is expected to provide theoretical support for elucidating the pathogenesis of HPH and discover potential therapeutic target genes of HPH.
Materials and methods
Data retrieval
GEO is a public functional genomics data repository. GSE154959 [16] and GSE131425 were screened out from GEO. GSE154959 is a dataset containing the scRNA-seq data of lung ECs isolated from three controls and three PAH mice. GSE131425 contains bulk RNA-seq data of lung tissues from six controls and four hypoxia-induced PAH mice.
scRNA-seq data processing
The scRNA-seq data were analyzed using the “Seurat” package (version 4.0.4). Cell quality control was carried out based on the following criteria: (1) genes detected in < 5 cells were excluded; (2) cells with < 300 total detected genes were excluded; and (3) cells with ≥ 20% of mitochondria-expressed genes were excluded. The functions of “IntegrateData” and “NormalizeData” in the “Seurat” package were used for batch effect elimination and normalization between the six samples. Subsequently, the Uniform Manifold Approximation and Projection (UMAP) algorithm was used for dimensionality reduction of the 30 initial PCs, and cluster analysis was performed for all units. The cell clusters were subjected to cell type annotation according to the cell marker genes (Supplementary Table 1) from literature and the CellMarker database (http://xteam.xbio.top/CellMarker/).
Subpopulation identification of pulmonary ECs [17]
The subpopulation markers of ECs were identified based on previously reported literature [18, 19]. The “FindConservedMarkers” function in “Seurat” package was used to identify proliferating cells’ markers, and we provided the detailed data list of marker genes corresponding to proliferating cells (Supplementary Table 2). The signature of proliferating cells was calculated using the average expression of proliferating cells’ markers in GSE131425.
Trajectory analysis of arterial and proliferating cells
Trajectory analysis reconstructs the change process of cells over time by constructing the change trajectories between cells. To map the differentiation of cell subsets in ECs in HPH, we use the “plot_cell_trajectory” in “Monocle2” package to perform trajectory analysis. The “FindConservedMarkers” function in “Seurat” package was used to select the top 2,000 highly variable features (HVGs) for construction of trajectories. The “BEAM” function in Monocle2 was used to define branch-specific genes.
Differential analysis and functional enrichment analysis
Relevant DEGs were screened using the “DESeq2” package. Heatmap of the DEGs was drawn using the “pheatmap” package. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses [20,21,22] were performed using the “clusterProfiler” package. DEGs with padj < 0.05 and |log2FC| > 1 were considered statistically significant.
Weighted gene co-expression network analysis (WGCNA)
WGCNA is designed to show the co-expression relationships between genes. It is a systems biological approach to describe gene association patterns between different samples. WGCNA can be used to identify gene sets with high covariation, and to identify candidate biomarker genes or therapeutic targets based on the endogeneity of gene sets and the association between gene sets and phenotypes. We calculated the correlation value of gene expression in GSE131425, and carried out suborder operation. The soft threshold was adjusted to 8 (scale free R2 = 0.8) to construct a scale-free network. R2 = 8 was used to construct an adjacency matrix of the differential genes, which was then transformed into a topological overlap matrix (TOM). Hierarchical clustering was then performed for module identification. Finally, the feature genes were calculated, the modules were hierarchically clustered, and similar modules were merged. 7 modules were subsequently identified.
Cell model preparation and grouping
An in vitro cell model was built by treating hPAECs with hypoxia to simulate HPH. The over-expression vector of Hpgd (OE-Hpgd) was constructed and transfected into hPAECs. At the same time, the empty plasmid (OE-NC) was transferred into hPAECs as a control. The transfected hPAECs were treated with hypoxia for 24 h, and were divided into hypoxia + OE-Hpgd group and hypoxia + OE-NC group.
Cell culture
The hPAECs (#CP-H002) used in this study were purchased from Pricella, inoculated in the complete culture medium for hPAECs (Pricella, #CM-H002), and cultured in an incubator with 5% CO2 at 37℃.
Western blot
The total protein was extracted from the cell lysate containing 1% protease inhibitor, and the protein in the samples was quantified by BCA method, electrophoresis, membrane transfer, and antibody incubation. The primary antibodies were Hpgd (Abcam, #ab187160), Bax (Abcam, #ab32503) and Bcl-2 (Abcam, #ab32124). GAPDH (Abcam, #ab8245) was selected as the internal reference gene. The optimum concentration was 0.418 µg/ml for Hpgd, 0.11 µg/ml for Bax, 0.4 µg/ml for Bcl-2, and 2 µg/ml for GAPDH. The protein strips were exposed and analyzed using Image J software.
qRT-PCR
Total RNA was extracted using the TRIzol kit (ThermoFisher, USA). The cDNA was obtained using the PrimeScript RT kit (TaKaRa, Japan), and then the cDNA template was diluted with RNase-free water. qPCR was performed according to the instructions of SYBR Green PCR kit (ThermoFisher, USA). The total reaction system was 10 µl. The condition of qRT-PCR: pre-denaturation at 95℃ for 15 min, denaturation at 94℃ for 15s, annealing at 55℃ for 30s, extension at 72℃ for 30s, and a total of 40 cycles. The primer sequences are as follows: Hpgd-forward primer (F): GCAAGCGAACCATTCCACTCTTTG; Hpgd-reverse primer (R): ACAGTTTCACATTCCCACCCATCAG; GAPDH-F: GATCATCAGCAATGCCTCCT; GAPDH-R: TGTGGTCATGAGTCCTTCCA. The qRT-PCR (#A28139) instrument was purchased from Thermo Fisher.
Assessment of the proliferation and apoptosis of hPAECs
According to the manufacturer’s instructions, the levels of proliferation and apoptosis of hPAECs were evaluated using Bromodeoxyuridine (BrdU) incorporation assay (spbio, China) and in situ Brdu-red DNA fragmentation (TUNEL) apoptosis assay kit (Beyotime, China), and the degree of apoptosis was evaluated by Bax/Bcl-2 ratio.
Adhesion and angiogenesis assay of the ECs
Adhesion experiments of ECs were performed according to the instructions of the cell adhesion kit (AAT Bioquest, 23,010). Additionally, the pre-cooled Matrigel matrix glue was added into the 24-well plates, and 500 µl of the cell suspension was added into each well. The results of angiogenesis assay were observed after routine culture in the cell incubator for 2-6 h. Image J and NIS-Elements BR Analysis (Nikon) were used to quantify adherent cells and tube formation.
Statistical analysis
The experimental data were processed using R language and GraphPad Prism 8.0. P > 0.05 denotes statistically significant difference.
Results
Differential analysis
The GSE131425 dataset were subjected to DEGs analysis. Based on P < 0.05 and |log2FC| > 1, 454 DEGs were identified, of which 277 were up-regulated and 277 were down-regulated (Fig. 1& Supplementary Table 3).
Heterogeneity of the ECs
The Seurat object contained 28,940 cells and 16,980 genes after quality control and batch correction. These cells were clustered into 21 clusters. According to the expression of cell marker genes in clusters obtained from literature review and CellMarker database, cell type annotations were made for the clusters (Fig. 2A). The ECs accounted for the largest proportion of all the samples (Fig. 2B). 6 subpopulations of the ECs were revealed through further analysis, and they were capillary A, capillary B, venous, arterial, lymphatic and proliferating cells (Fig. 2C). 183 proliferating cell markers were identified with the aid of “FindConservedMarkers” function.
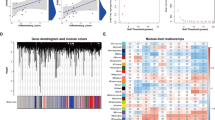
WGCNA
Pearson’s correlation coefficient was used to cluster the samples, and a sample clustering tree was drawn correspondingly (Fig. 3A). The soft threshold was adjusted to 8 (R2 = 0.8) for scale-free network construction (Fig. 3B). Next, the adjacency matrix was built and the TOM was constructed. Finally, 7 modules were identified based on average hierarchical clustering and dynamic tree clipping (Fig. 3C). The correlation analysis of different modules and indicators (HPH and the proliferating cell signature) showed that the blue module was highly correlated with HPH and the proliferating cell signature (Fig. 3D, E). Therefore, it was chosen as the clinically important module for follow-up analysis. The blue module contained 181 genes, and 41 key genes were screened out according to Gene Significance (GS) > 0.9 and Module Membership (MM) > 0.9. GO and KEGG enrichment analysis was performed on the key genes (Fig. 4A, B). Most of these genes were shown to be enriched in muscle cell proliferation, peroxisome, cytokine-cytokine receptor interaction, and other functions.
Trajectory analysis
The trajectory analysis projects all arterial cells and proliferating cells onto five states. The results showed that the cells ranged from proliferating cells to arterial cells in the pseudo-time line, and the proliferating cells were mostly from the HPH group (Fig. 5A-C). According to cell differentiation, cell was divided into five states (Fig. 5D). Branch point 1 was analyzed using BEAM method and the results were clustered (threshold, qval < 1e-6). These genes can be clustered into 5 categories (Fig. 5E), where pre-branch represents state 3, cell fate 1 represents state 4, and cell fate 2 represents state 5. Cluster 1 contains a large number of genes, and the overall trend is roughly consistent, so we choose cluster 1 for subsequent analysis. Cluster 1 contains 523 genes. GO and KEGG enrichment analysis were performed on the genes in cluster 1 (Fig. 5F, G), and the results showed that most of the genes were enriched in the regulation of vasculature development, endocytosis, phagosome and other related functions.
Hub genes
Three key genes (Hpgd, Npr3 and Fbln2) were finally screened out through intersection between the key genes of the blue module and the genes in cluster 1. Compared with the control group, Hpgd and Npr3 were down-regulated, and Fbln2 was up-regulated in PAH (Fig. 6A). The expression level of Hpgd in ECs was much higher than that of Npr3 and Fbln2 (Fig. 6B). In addition, through trajectory analysis, Hpgd showed an obvious trend of change in the process of cell differentiation (Fig. 6C), so Hpgd was selected for cell experiment verification.
Hpgd expression decreased in hypoxia-treated hPAECs
hPAECs were treated with different durations of hypoxia (0, 6, 12, 24, 48 h) in order to simulate HPH cell model in vitro. Western blot and qRT-PCR were used to detect the expression of Hpgd in hPAECs treated with different durations of hypoxia. Hpgd expression decreased in a time-dependent manner in hypoxia-treated hPAECs (Fig. 7A, B). This result was consistent with the results of bioinformatics analysis above.
The expression levels of Hpgd in hPAECs treated with hypoxia for 0, 6, 12, 24 and 48 h. (A) Western Blot results showed that the expression of Hpgd decreased in hypoxia-treated hPAECs. (B) qRT-PCR showed that the longer the time of hypoxic treatment of hPAECs, the lower the expression of Hpgd. (*P<0.05, **P<0.01)
Effects of Hpgd on the proliferation and apoptosis of hypoxia-treated hPAECs
It is known that the expression of Hpgd decreased in hypoxia-treated hPAECs. In order to study the effect of Hpgd on the proliferation and apoptosis of hPAECs, OE-NC/OE-Hpgd was transfected into hPAECs (Fig. 8A). The proliferation activity of hPAECs was detected by BrdU incorporation assay, and the apoptosis was detected by TUNEL staining. It was found that compared with the hypoxia + OE-NC group, the proliferation activity in the hypoxia + OE-Hpgd group decreased, and the number of apoptotic cells increased (Fig. 8B-D). Western blot was used to detect the expression of apoptosis-related proteins (Bax and Bcl2). Compared with the hypoxia + OE-NC group, the Bax/Bcl2 ratio in the hypoxia + OE-Hpgd group increased (Fig. 8E, F). These results indicated that Hpgd regulated the proliferative activity and apoptosis of hypoxia-treated hPAECs.
Effects of Hpgd on the proliferation and apoptosis of hPAECs. (A) Western blot verified that hPAECs was successfully transfected with OE-Hpgd. (B) BrdU incorporation assay showed that OE-Hpgd inhibited cell proliferation. (C) TUNEL staining results. (D) TUNEL staining showed that OE-Hpgd increased the number of apoptotic cells. (E) The expression of apoptosis-related proteins shown by Western blot. (F) The ratio of Bax/Bcl-2 expression showed that the degree of apoptosis was deepened. (*P<0.05, **P<0.01)
Effects of Hpgd on the adhesion and angiogenesis of hypoxia-treated hPAECs
In order to investigate the effect of Hpgd on the adhesion and angiogenesis of hPAECs, the levels of adhesion and angiogenesis of hPAECs transfected with OE-NC/OE-Hpgd were tested. It was found that both adherent cells and angiogenesis (number of tubules) were reduced in the hypoxia + OE-Hpgd group when compared with the hypoxia + OE-NC group (Fig. 9A-D), indicating that Hpgd regulated the adhesion and angiogenesis of hypoxia-treated hPAECs.
Effects of Hpgd on the adhesion and angiogenesis of hPAECs. (A) The microscope results showed that OE-Hpgd reduced the adhesion of hPAECs. (B) Statistical histogram of the adherent cells. (C) The microscope results showed that OE-Hpgd reduced the angiogenesis capability. (D) Statistical bar chart of the number of tubules. (**P<0.01)
Discussion
HPH is caused by hypoxia-induced pulmonary vasoconstriction, thus increasing pulmonary artery pressure. At present, due to the limited detection methods, HPH is less liable to be detected in time. Moreover, HPH is mostly induced by basic pulmonary diseases that complicate the course of chronic diseases [23]. Current treatments for HPH generally give priority to the underlying lung diseases [24, 25]. However, the pathogenesis of HPH remains unclear.
In order to investigate the mechanism of HPH, we downloaded the scRNA-seq dataset GSE154959 and the bulk RNA-seq dataset GSE131425 from GEO. According to the identification of cell types by using the scRNA-seq data, it was found that ECs accounted for the largest proportion and had six subtypes: capillary A, capillary B, venous, arterial, lymphatic, proliferating cells that suggested the heterogeneity of ECs. Recent scRNA-seq profiles of mouse ECs show that ECs are heterogeneous in the brain, muscles, heart, kidney and other tissues [26, 27]. The heterogeneity of ECs has been established early in the embryonic development, and is gradually adjusted and enhanced with each developmental stage. At the same time, such heterogeneity may also change under different pathological conditions [28]. According to existing studies, ECs proliferate abnormally with the occurrence of HPH, causing vascular remodeling [29]. Through trajectory analysis of arterial cells and proliferating cells, 523 key genes were identified, and they were revealed to be enriched in the regulation of vasculature development, endocytosis, phagosome by GO and KEGG analysis. To some extent, key genes associated with the transformation from proliferating cells to arterial cells were considered relevant to vascular remodeling. Piperlongumine has been shown to treat HPH by regulating autophagy and reducing vascular remodeling [30]. Fasudil is known to interfere with vascular remodeling, reduce proliferation and apoptosis of pulmonary ECs, and alleviate the occurrence of HPH [31]. Galectin-3, expressed in vascular SMCs, promotes PAH by regulating changes in the proliferation, apoptosis, and fibrosis of the cells [32]. Nicotinic acid can inhibit vascular remodeling by releasing H-PGDS from macrophages and promoting the production of PGD2 in lung tissues, thus improving the progression of PAH [33]. T cells are regulated to up-regulate pulmonary and right ventricular vascular protective protein COX-2 (cyclooxygenase 2) in ECs to counteract the pulmonary vascular damage produced in HPH [34].
WGCNA of the bulk RNA-seq data was performed to identify important clinical modules, and 41 key genes were screened out based on GS > 0.9 and MM > 0.9. Most of these genes were shown by GO and KEGG enrichment analysis to be enriched in the regulation of vascular development, SMC proliferation and immunity-related functions. 3 key genes (Hpgd, Npr3 and Fbln2) were obtained through intersection of genes screened from the two datasets mentioned above, and their expressions were analyzed in the meantime. Compared with the control group, Hpgd and Npr3 were down-regulated in PAH, and Fbln2 was up-regulated in PAH.
Natriuretic peptide receptor 3 (Npr3) encodes a receptor clearance gene for final degradation by binding and internalizing natriuretic peptides (NPs). It protects cardiomyocytes from apoptosis through inhibition of cytosolic BRCA1 and TNF-α, which are regulators of apoptosis [35]. Fibulin-2 (Fbln2) codes for a gene that secretes an extracellular matrix glycoprotein and is involved in tissue development and remodeling. Fbln2 is expressed in the epithelial-mesenchymal transition of the central endometrial cushion matrix during embryonic heart development [36]. According to Shaukat’s study, the expression of Fbln2 is a key regulator of angiotensin II-induced TGF-β signaling and subsequent myocardial fibrosis [37]. Hpgd (15-hydroxyprostaglandin dehydrogenase) encodes a member of the short chain nonmetallic enzyme alcohol dehydrogenase protein family, and can activate STAT3 and AKT pathways to promote proliferation, migration and anchorage-independent growth of cervical cancer cells [38]. Deletion of Hpgd in regulatory T cells (Treg cells) in mice leads to abnormal proliferation and accumulation of functionally impaired Treg cells, inducing local inflammation [39]. miR-106b-5p regulates the progression of esophageal squamous cell carcinoma by binding to Hpgd [40]. However, there has been no corresponding study on Hpgd in HPH. Therefore, we verified the role of Hpgd by in vitro experiments. After hypoxic treatment of hPAECs, the expression of Hpgd was decreased in a time-dependent pattern, which was consistent with the results drawn from bioinformatics analysis. Hpgd overexpression was revealed to regulate the proliferative activity, apoptosis level, adhesion and angiogenesis capability of hypoxia-treated hPAECs.
Conclusion
In this study, bioinformatics analysis was carried out based on both of the scRNA-seq data and bulk RNA-seq data, through which we found that Hpgd may play a key role in the occurrence and development of HPH. Through cellular experimental verification, Hpgd was shown to decrease the proliferation capacity, increase the level of apoptosis, and reduce the adhesion and angiogenesis ability of ECs. This study deepens the current understanding of the pathogenesis of HPH, which may enable us to conduct intervention therapy for HPH targeting Hpgd.
Data Availability
The datasets used and/or analysed during the current study are available from the GEO database (http://www.ncbi.nlm.nih.gov/geo) [Accession number: GSE154959 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154959) & Accession number: GSE131425 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131425)].
Abbreviations
- HPH:
-
Hypoxic pulmonary hypertension
- scRNA-seq:
-
single cell sequencing
- RNA-seq:
-
RNA sequencing
- GEO:
-
Gene Expression Omnibus
- WGCNA:
-
weighted correlation network analysis
- hPAECs:
-
human pulmonary artery endothelial cells
- ECs:
-
endothelial cells
- SMCs:
-
smooth muscle cells
- COPD:
-
chronic obstructive pulmonary disease
- PAH:
-
Pulmonary arterial hypertension
- WHO:
-
World Health Organization
- DEGs:
-
differentially expressed genes
- UMAP:
-
Uniform Manifold Approximation and Projection
- HVGs:
-
highly variable features
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- TOM:
-
topological overlap matrix
- BrdU:
-
Bromodeoxyuridine
- GS:
-
Gene Significance
- MM:
-
Module Membership
- Npr3:
-
Natriuretic peptide receptor 3
- Fbln2:
-
Fibulin-2
References
Woo KV, Ornitz DM, Singh GK. Diagnosis and pathophysiological mechanisms of Group 3 Hypoxia-Induced Pulmonary Hypertension. Curr Treat Options Cardiovasc Med. 2019;21(3):16.
Poch D, Mandel J. Pulmonary hypertension. Ann Intern Med. 2021;174(4):ITC49–ITC64.
Zhao Y, Wang B, Zhang J, He D, Zhang Q, Pan C, et al. ALDH2 (Aldehyde dehydrogenase 2) protects against Hypoxia-Induced Pulmonary Hypertension. Arterioscler Thromb Vasc Biol. 2019;39(11):2303–19.
Yun S, Junbao D, Limin G, Chaomei Z, Xiuying T, Chaoshu T. The regulating effect of heme oxygenase/carbon monoxide on hypoxic pulmonary vascular structural remodeling. Biochem Biophys Res Commun. 2003;306(2):523–9.
Xu D, Li Y, Zhang B, Wang Y, Liu Y, Luo Y, et al. Resveratrol alleviate hypoxic pulmonary hypertension via anti-inflammation and anti-oxidant pathways in rats. Int J Med Sci. 2016;13(12):942–54.
Vonk Noordegraaf A, Westerhof BE, Westerhof N. The Relationship between the right ventricle and its load in Pulmonary Hypertension. J Am Coll Cardiol. 2017;69(2):236–43.
Rowan SC, Keane MP, Gaine S, McLoughlin P. Hypoxic pulmonary hypertension in chronic lung diseases: novel vasoconstrictor pathways. Lancet Respir Med. 2016;4(3):225–36.
Hoeper MM, McLaughlin VV, Dalaan AM, Satoh T, Galie N. Treatment of pulmonary hypertension. Lancet Respir Med. 2016;4(4):323–36.
Liu J, Wang W, Wang L, Chen S, Tian B, Huang K, et al. IL-33 initiates vascular remodelling in hypoxic pulmonary hypertension by up-regulating HIF-1alpha and VEGF expression in vascular endothelial cells. EBioMedicine. 2018;33:196–210.
Dai Z, Zhu MM, Peng Y, Jin H, Machireddy N, Qian Z, et al. Endothelial and smooth muscle cell Interaction via FoxM1 Signaling mediates vascular remodeling and pulmonary hypertension. Am J Respir Crit Care Med. 2018;198(6):788–802.
Wang Q, Tian J, Li X, Liu X, Zheng T, Zhao Y, et al. Upregulation of endothelial DKK1 (dickkopf 1) promotes the development of Pulmonary Hypertension through the Sp1 (specificity protein 1)/SHMT2 (serine hydroxymethyltransferase 2) pathway. Hypertension. 2022;79(5):960–73.
Aird WC. Endothelial cell heterogeneity. Cold Spring Harb Perspect Med. 2012;2(1):a006429.
Hennigs JK, Matuszcak C, Trepel M, Korbelin J. Vascular Endothelial Cells: Heterogeneity and Targeting Approaches.Cells. 2021;10(10).
Brown CC, Gudjonson H, Pritykin Y, Deep D, Lavallee VP, Mendoza A, et al. Transcriptional basis of mouse and human dendritic cell heterogeneity. Cell. 2019;179(4):846–63. e24.
Liang L, Yu J, Li J, Li N, Liu J, Xiu L, et al. Integration of scRNA-Seq and bulk RNA-Seq to Analyse the heterogeneity of Ovarian Cancer Immune cells and establish a molecular risk model. Front Oncol. 2021;11:711020.
Rodor J, Chen SH, Scanlon JP, Monteiro JP, Caudrillier A, Sweta S, et al. Single-cell RNA sequencing profiling of mouse endothelial cells in response to pulmonary arterial hypertension. Cardiovasc Res. 2022;118(11):2519–34.
Schupp JC, Adams TS, Cosme C Jr, Raredon MSB, Yuan Y, Omote N, et al. Integrated single-cell atlas of endothelial cells of the human lung. Circulation. 2021;144(4):286–302.
Muller AM, Hermanns MI, Skrzynski C, Nesslinger M, Muller KM, Kirkpatrick CJ. Expression of the endothelial markers PECAM-1, vWf, and CD34 in vivo and in vitro. Exp Mol Pathol. 2002;72(3):221–9.
Vicen M, Igreja Sa IC, Tripska K, Vitverova B, Najmanova I, Eissazadeh S, et al. Membrane and soluble endoglin role in cardiovascular and metabolic disorders related to metabolic syndrome. Cell Mol Life Sci. 2021;78(6):2405–18.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587–D92.
Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–51.
Nathan SD, Barbera JA, Gaine SP, Harari S, Martinez FJ, Olschewski H et al. Pulmonary hypertension in chronic lung disease and hypoxia.Eur Respir J. 2019;53(1).
Sakao S. Chronic obstructive pulmonary disease and the early stage of cor pulmonale: a perspective in treatment with pulmonary arterial hypertension-approved drugs. Respir Investig. 2019;57(4):325–9.
Wu G, Lee YY, Gulla EM, Potter A, Kitzmiller J, Ruben MD et al. Short-term exposure to intermittent hypoxia leads to changes in gene expression seen in chronic pulmonary disease.Elife. 2021;10.
Kalucka J, de Rooij L, Goveia J, Rohlenova K, Dumas SJ, Meta E, et al. Single-cell transcriptome atlas of murine endothelial cells. Cell. 2020;180(4):764–79. e20.
Vila Ellis L, Cain MP, Hutchison V, Flodby P, Crandall ED, Borok Z, et al. Epithelial Vegfa specifies a distinct Endothelial Population in the mouse lung. Dev Cell. 2020;52(5):617–30. e6.
Guo FH, Guan YN, Guo JJ, Zhang LJ, Qiu JJ, Ji Y, et al. Single-cell transcriptome analysis reveals embryonic endothelial heterogeneity at Spatiotemporal Level and Multifunctions of MicroRNA-126 in mice. Arterioscler Thromb Vasc Biol. 2022;42(3):326–42.
Masri FA, Xu W, Comhair SA, Asosingh K, Koo M, Vasanji A, et al. Hyperproliferative apoptosis-resistant endothelial cells in idiopathic pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol. 2007;293(3):L548–54.
Ye W, Tang T, Li Z, Li X, Huang Q. Piperlongumine attenuates vascular remodeling in hypoxic pulmonary hypertension by regulating autophagy. J Cardiol. 2022;79(1):134–43.
Sun XZ, Tian XY, Wang DW, Li J. Effects of fasudil on hypoxic pulmonary hypertension and pulmonary vascular remodeling in rats. Eur Rev Med Pharmacol Sci. 2014;18(7):959–64.
Barman SA, Li X, Haigh S, Kondrikov D, Mahboubi K, Bordan Z, et al. Galectin-3 is expressed in vascular smooth muscle cells and promotes pulmonary hypertension through changes in proliferation, apoptosis, and fibrosis. Am J Physiol Lung Cell Mol Physiol. 2019;316(5):L784–L97.
Jia D, Bai P, Wan N, Liu J, Zhu Q, He Y, et al. Niacin attenuates pulmonary hypertension through H-PGDS in macrophages. Circ Res. 2020;127(10):1323–36.
Tamosiuniene R, Manouvakhova O, Mesange P, Saito T, Qian J, Sanyal M, et al. Dominant Role for Regulatory T cells in protecting females against pulmonary hypertension. Circ Res. 2018;122(12):1689–702.
Lin D, Chai Y, Izadpanah R, Braun SE, Alt E. NPR3 protects cardiomyocytes from apoptosis through inhibition of cytosolic BRCA1 and TNF-alpha. Cell Cycle. 2016;15(18):2414–9.
Tsuda T, Wang H, Timpl R, Chu ML. Fibulin-2 expression marks transformed mesenchymal cells in developing cardiac valves, aortic arch vessels, and coronary vessels. Dev Dyn. 2001;222(1):89–100.
Khan SA, Dong H, Joyce J, Sasaki T, Chu ML, Tsuda T. Fibulin-2 is essential for angiotensin II-induced myocardial fibrosis mediated by transforming growth factor (TGF)-beta. Lab Invest. 2016;96(7):773–83.
Yao S, Xu J, Zhao K, Song P, Yan Q, Fan W, et al. Down-regulation of HPGD by miR-146b-3p promotes cervical cancer cell proliferation, migration and anchorage-independent growth through activation of STAT3 and AKT pathways. Cell Death Dis. 2018;9(11):1055.
Schmidleithner L, Thabet Y, Schonfeld E, Kohne M, Sommer D, Abdullah Z, et al. Enzymatic activity of HPGD in Treg cells suppresses Tconv cells to maintain adipose tissue homeostasis and prevent metabolic dysfunction. Immunity. 2019;50(5):1232–48e14.
Yang F, Sun Z, Wang D, Du T. MiR-106b-5p regulates esophageal squamous cell carcinoma progression by binding to HPGD. BMC Cancer. 2022;22(1):308.
Acknowledgements
Not applicable.
Funding
This work was supported by the Natural Science Foundation of Zhejiang Province [Grant No. LQ20H010001], Zhejiang Traditional Chinese Medicine Science and Technology Program [Grant No. 2023ZL183], Shaoxing Health Technology Program [Grant No. 2022KY006], and Shaoxing Medical and Health Science and Technology Project [Grant No. 2020A13020].
Author information
Authors and Affiliations
Contributions
MH and KL-T originated the research concept and designed the study. MX and JS collected and assembled the data. MH, KL-T and MX performed data analysis. MH wrote the first draft of the manuscript. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
He, M., Tao, K., Xiang, M. et al. Hpgd affects the progression of hypoxic pulmonary hypertension by regulating vascular remodeling. BMC Pulm Med 23, 116 (2023). https://doi.org/10.1186/s12890-023-02401-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12890-023-02401-y