Abstract
Background
This study analysed the performance of radiomics features extracted from computed tomography (CT) images with different reconstruction parameters in differentiating malignant and benign pulmonary nodules.
Methods
We evaluated routine chest CT images acquired from 148 participants with pulmonary nodules, which were pathologically diagnosed during surgery in West China Hospital, including a 5 mm unenhanced lung window, a 5 mm unenhanced mediastinal window, a 5 mm contrast-enhanced mediastinal window and a 1 mm unenhanced lung window. The pulmonary nodules were segmented, and 1409 radiomics features were extracted for each window. Then, we created 15 cohorts consisting of single windows or multiple windows. Univariate correlation analysis and principal component analysis were performed to select the features, and logistic regression analysis was performed to establish models for each cohort. The area under the curve (AUC) was applied to compare model performance.
Results
There were 75 benign and 73 malignant pulmonary nodules, with mean diameters of 18.63 and 19.86 mm, respectively. For the single-window setting, the AUCs of the radiomics model from the 5 mm unenhanced lung window, 5 mm unenhanced mediastinal window, 5 mm contrast-enhanced mediastinal window and 1 mm unenhanced lung window were 0.771, 0.808, 0.750, and 0.771 in the training set and 0.711, 0.709, 0.684, and 0.674 in the test set, respectively. Regarding the multiple-window setting, the radiomics model based on all four windows showed an AUC of 0.825 in the training set and 0.743 in the test set. Statistically, the 15 models demonstrated comparable performances (P > 0.05).
Conclusion
A single chest CT window was acceptable in predicting the malignancy of pulmonary nodules, and additional windows did not statistically improve the performance of the radiomics models. In addition, slice thickness and contrast enhancement did not affect the diagnostic performance.
Similar content being viewed by others
Introduction
Pulmonary nodules are commonly detected on computed tomography (CT) of the chest [1]. About 95% of detected pulmonary nodules are benign and have a wide variety of causes (most often granulomas or intrapulmonary lymph nodes), while a small number of nodules are early lung cancers [2, 3]. The optimal diagnostic approach for pulmonary nodules should facilitate timely and effective curative treatment for lung cancer and simultaneously avoid harmful interventions in benign disease [1]. At present, most guidelines recommend noninvasive risk assessment of pulmonary nodules; and CT surveillance is recommended for low-risk nodules, while PET-CT or biopsy or excision is suggested for high-risk nodules [1, 4, 5]. The recommended risk assessment models mainly include the Brock model and Mayo model, which were established based on clinical variables [6, 7].
Nevertheless, drastic increases in computational power and memory have enabled the development and implementation of artificial intelligence techniques in the medical field [8]. For example, radiomics analysis, which can noninvasively mine high-throughput quantitative image features from standard-of-care medical imaging, is gaining incremental importance in cancer research [9]. Most studies have demonstrated that radiomics models could be effective supplementary tools for the decision-making of clinicians in the diagnosis and treatment of cancer, and radiomics models can predict the malignancy of lesions, gene mutation, pathological type, clinical stage, treatment response and prognosis [10]. For pulmonary nodules, previous studies have shown that radiomics models performed well in predicting the lung cancer risk of nodules based on CT images, such as low-dose or routine CT images [11, 12] and unenhanced or contrast-enhanced CT images [13, 14].
However, it was reported that image acquisition and reconstruction parameters such as contrast enhancement, slice thickness, convolution algorithm, tube voltage and current could affect the reproducibility of radiomic features and influence the diagnostic performance of radiomics models [15,16,17]. For example, Stefano et al. revealed that the diagnostic value of histogram features could vary at different Hounsfield units (HU) [18]. Nevertheless, only one or a few reconstruction parameters were investigated in previous studies of lung cancer risk prediction. The impact of different reconstruction parameters on radiomics feature extraction and analysis has not yet been thoroughly explored.
Hence, this study intended to simultaneously assess how four common chest CT reconstruction windows affect the performance of radiomics features in differentiating malignant and benign pulmonary nodules, which included a 5 mm unenhanced lung window, a 5 mm unenhanced mediastinal window, a 5 mm contrast-enhanced mediastinal window and a 1 mm unenhanced lung window.
Methods
Study patients
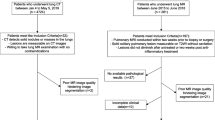
This retrospective, single-centre study was approved by the institutional review board of the West China Hospital of Sichuan University, and the requirement for informed consent was waived as the privacy and identity information of the participants were protected. The study selected eligible patients discharged from West China Hospital of Sichuan University from 2010 to 2018 based on the following inclusion criteria: (1) the patient was older than 18 years old; (2) there was a solitary pulmonary nodule detected on routine chest CT in the institution; (3) the patient was treated with surgery without receiving chemo- or radiotherapy, and the nodule was pathologically confirmed as primary lung cancer or benign pulmonary lesion; and (4) four chest CT reconstruction windows were available, including a 5 mm unenhanced lung window, a 5 mm unenhanced mediastinal window, a 5 mm contrast-enhanced mediastinal window and a 1 mm unenhanced lung window. Otherwise, the subjects were excluded if (1) the patient was receiving treatment due to other malignancies; (2) the pulmonary nodule was calcified; or (3) the entire nodule volume was not completely shown on all four windows. As the 1 mm contrast-enhanced mediastinal window was only available in a small number of patents, this window was not collected and analysed in the current study. In total, 148 patients with 592 chest CT images were finally enrolled. Clinical variables, including sex, age, and pathological diagnosis, were collected for further analysis.
CT acquisition protocol
Images were acquired from one 64-slice multidetector CT scanner of the chest (SOMATOM, Definition Flash, Siemens). The scan parameters included: voltage, 80–140 kVp; current, 98–678 mAs; rotation time, 0.5 s; matrix, 512 × 512; table speed, 30.6-139.3 mm/s; reconstruction thickness, 1 and 5 mm. After plain CT, arterial-phase enhanced scans were performed 40 s after intravenous injection of nonionic iodinated contrast medium (300 mg/mL), which was administered for all patients at a dose of 2 mL/kg body weight and rate of 3.5-4.0 mL/s using a power injector from Bayer. Finally, four CT series were reconstructed: 5 mm unenhanced lung window, 5 mm unenhanced mediastinal window, 5 mm contrast-enhanced mediastinal window and 1 mm unenhanced lung window. All images were exported in DICOM format for subsequent analysis.
CT analysis
Two respiratory physicians (CBJ and ZR, with 10 and 6 years of experience in chest image interpretation, respectively) independently reviewed all the CT images and resolved discrepancies by discussion. All images were reviewed at both lung (width = 1500 HU; level = − 700 HU) and mediastinal (width = 350 HU; level = 40 HU) settings. Nodule diameter, location, texture, consolidation/tumour ratio, spiculation, lobulation and cavity were evaluated on the axial plane of a 1 mm unenhanced lung window as morphological features. The consolidation/tumour ratio, namely, the ratio of the maximum consolidation diameter to the maximum nodule diameter, was calculated to assess the proportion of the solid component [19].
Image preprocessing, nodule segmentation and radiomics feature extraction
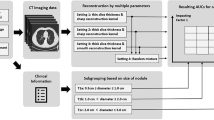
Figure 1 shows the whole modelling pipeline. First, LK software (Lung Intelligence Kit, version 1.5.0, GE Healthcare) was applied to preprocess images and segment the volume of interest (VOI). All images were preprocessed by resampling (spatial resolution = 1 mm × 1 mm × 1 mm), Laplace enhancement and Gaussian filtering (standard deviation, SD = 0.5). Then, the LK software automatically detected and segmented the whole nodule volume on each reconstruction window, which was accompanied by manual correction (by CBJ and ZR) to ensure that every nodule was accurately delineated. Approximately 40% of the nodules segmented by the software were manually modified slice by slice. One example of a segmented nodule on four windows is shown in Fig. 2.
Subsequently, AK software (Artificial Intelligence Kit, version 3.3.0, GE Healthcare) was applied to automatically extract 1409 three-dimensional radiomics features from each window for all patients, including shape features (n = 14), first-order features (n = 18), grey level cooccurrence matrix features (n = 24), grey level run length matrix features (n = 16), grey level size zone matrix features (n = 16), neighbouring grey tone difference matrix features (n = 5), grey level dependence matrix features (n = 14), Laplacian of Gaussian filtered features (n = 186, Sigma = 1.0, 3.0, 5.0), first-order wavelet-filtered features (n = 744, Level = 1) and 3-D local binary pattern related features (n = 372, Level = 2, Radius = 1.00, Subdivision = 1).
Feature selection and model construction
To evaluate the diagnostic value of radiomics features derived from independent reconstruction windows as well as multiple reconstruction windows, we regrouped the four windows into 15 cohorts to establish 15 models, including four single-window models, six double-window models, four triple-window models and one four-window model. All subjects were randomly separated into the training set (N = 104) and test set (N = 44) at a ratio of 7:3.
As there were thousands of radiomics features in each cohort, feature selection was performed to identify representative features and avoid overfitting. Specifically, after standardization with Z score transformation, univariate correlation analysis (cut-off = 0.7) and principal component analysis (number of components = 10) were performed to select features. In the study, the number of principal components was set to ten based on the scree diagram derived from Kaiser‒Meyer‒Olkin analysis. Finally, the logistic regression model was selected and established in the training set and validated in the test set for each cohort after comparison with the other machine learning models (support vector machine, decision tree, decision forest, Bayes, K-nearest neighbour).
Statistical analysis
The continuous variables are described as the mean ± SD and were compared with Student’s t test, while the categorical variables are described as the number of cases (proportion) and were compared with the chi-square test or Fisher’s exact test. In addition, the receiver operating characteristic (ROC) curve and area under the curve (AUC) were acquired to evaluate the discrimination performance of the models. Delong’s test was conducted to compare AUCs. The accuracy, sensitivity and specificity were calculated based on Youden’s J index. Moreover, the Hosmer‒Lemeshow test was applied to illustrate the goodness of fit, and decision curve analysis was conducted to evaluate the clinical usefulness of the established models. A two-tailed p value < 0.05 indicated statistical significance. All statistical analyses were implemented using the Institute of Precision Medicine Statistics (version 1.1, GE Healthcare) and SPSS (version 28.0).
Results
Clinical characteristics
The clinical characteristics of the enrolled patients are summarized in Table 1. Among all 148 patients, the age ranged from 26 to 77 years old, with a mean ± SD of 54.49 ± 11.1 years. The patients with malignant nodules were older (57.12 ± 10.56 vs. 51.92 ± 11.05 years, P = 0.004). There were 57 males and 91 females, and no significant difference was observed between the malignant and benign groups (P = 0.293).
Regarding the pathology of the enrolled subjects, 75 (50.68%) nodules were benign, and 73 (49.32%) nodules were malignant. The malignant nodules included adenocarcinomas (N = 68, 93.15%) and squamous carcinomas (N = 5, 6.85%), whereas the benign nodules consisted of chronic inflammatory lesions (N = 30, 40.00%), granulomas (N = 15, 20.00%), hamartomas (N = 11, 14.67%), tuberculosis (N = 8, 10.67%) and so on. Detailed information on the pathological diagnosis of the pulmonary nodules is summarized in Table S1.
In terms of the morphological features of the enrolled malignant and benign nodules, the mean diameters were 19.15 mm and 18.63 mm, respectively (P = 0.672), and most were located in the right upper lobe (36.99% vs. 32.00%, P = 0.953). There were many more subsolid nodules in the malignant group (69.86% vs. 24.00%, P < 0.001). However, no significant difference was observed regarding spiculation (28.77% vs. 32.00%, P = 0.669), lobulation (31.51% vs. 29.33%, P = 0.774) and cavity sign (9.59% vs. 2.67%, P = 0.095) between the malignant and benign groups.
Radiomics feature selection
When the four windows were regrouped into 15 cohorts, there were 1409 features for the single-window cohort, 2818 features for the double-window cohort, 4227 features for the triple-window cohort and 5636 features for the four-window cohort. After univariate correlation analysis, approximately 10.8% ± 1% radiomics features remained in each cohort. Then, the remaining features were compressed to 10 representative components by principal component analysis to establish models for each cohort. Details of the radiomics feature selection are demonstrated in Table S2.
Performance of radiomics models
Figure 3 shows the ROC curves of the 15 established models, and Table 2 summarizes the corresponding diagnostic values. There were 11 models with an AUC greater than 0.700 in the test set. However, no significant differences were observed among the AUCs by DeLong’s tests (Table S3). Taken as a whole, all models demonstrated similar performance.
In detail, the four-window model demonstrated an AUC of 0.743 (95% CI, 0.607–0.862), an accuracy of 0.644, a sensitivity of 0.682 and a specificity of 0.609. For the single-window models, first, the 5 mm unenhanced-lung-window model showed an AUC of 0.711 (95% CI, 0.570, 0.840), an accuracy of 0.689, a sensitivity of 0.727 and a specificity of 0.652. Second, the 5 mm unenhanced mediastinal window model showed an AUC of 0.709 (95% CI, 0.567, 0.830), an accuracy of 0.644, a sensitivity of 0.636 and a specificity of 0.652. Third, the 5 mm contrast-enhanced mediastinal window model demonstrated an AUC of 0.684 (95% CI, 0.540, 0.814), an accuracy of 0.622, a sensitivity of 0.955 and a specificity of 0.304. Finally, the 1 mm unenhanced-lung-window model had an AUC of 0.674 (95% CI, 0.528, 0.802), an accuracy of 0.644, a sensitivity of 0.682 and a specificity of 0.609. The mentioned diagnostic values were from the test set.
As shown in Table 2, the values predicted by most models matched well with the actual data (Hosmer‒Lemeshow Test, P > 0.05). Figure 4 demonstrates the decision curve analyses for 15 models, which indicated that the established radiomics models had potential clinical practicability to some degree.
The decision curve analyses for all models in the training set (A) and test set (B). 5 L, 5 mm unenhanced lung window; 5 S, 5 mm unenhanced mediastinal window, 5 C, 5 mm contrast-enhanced mediastinal window; 1 mm, 1 mm unenhanced lung window; other model symbols being combinations of single-window abbreviations
Discussion
In the current study, by regrouping four chest CT reconstruction windows, a total of 15 single-window and multiple-window radiomics models were established. The results indicated that all models demonstrated similar performance in differentiating malignant and benign pulmonary nodules.
Some multiple-window radiomics models demonstrated higher AUCs than single-window models, but no significant differences were observed. Our results are consistent with those of Yang et al., who showed that radiomics signatures from both plain and vein-phase CT images were not superior to either plain signatures or vein signatures in differentiating solitary granulomas and solid lung adenocarcinomas [14]. However, two recent studies on lung cancer indicated that multiple-window-based models performed better than single-window-based models [20, 21]. For example, Lu et al. found that when predicting lung tumour growth patterns from radiomics features, the models constructed based on the lung window or the difference region (subtracting the mediastinal window region from the lung window region) were inferior to the model established based on both of them [21]. A possible explanation for these findings is that different strategies were applied in acquiring VOIs, and different kinds of features were used in the models.
In our investigations of diagnostic prediction, thinner reconstruction slice thickness did not provide added diagnostic information. Similarly, Park et al. found that the performance of radiomics models in prognostic prediction was not significantly changed when they were applied to 1-, 3-, and 5-mm slice thickness datasets [22]. Nevertheless, He et al. showed that thin-section CT-based radiomics signatures indeed had better diagnostic performance for solitary pulmonary nodules [23]. Our unexpected results may be explained by the larger nodule diameters of the enrolled patients (mean, 19.15 mm). As shown in one recent study, CT reconstruction parameters (including slice thickness) did not substantially affect the diagnostic performance of radiomics signatures for pulmonary nodules larger than 10 mm, but it mattered for pulmonary nodules less than 10 mm, where thin-section CT-based radiomics signatures performed better [24]. This study pointed out that for nodules greater than 10 mm, perhaps there is an optimal amount of imaging slices at which further addition will no longer increase diagnostic efficacy and may even decrease performance due to increased noise [24].
It is controversial how contrast enhancement affects radiomics analysis in predicting the malignancy of pulmonary nodules. He et al. showed that radiomics signatures from unenhanced CT demonstrated better discrimination and classification capability, as the biological heterogeneity within the tumour may be confounded by the contrast material [23]. On the other hand, Wu et al. found that contrast enhancement did not impact the utility of radiomics analysis, and the hypothesis was that the selected radiomics features measure the correlation, uniformity, and deviation of the pixels [13]. Nevertheless, our results indicated that radiomics signatures from the unenhanced and contrast-enhanced CT images had comparable performance in lung cancer risk prediction, where no significant difference was observed.
There were some limitations in the current study. First, this was a retrospective, single-centre study with a relatively small population. Second, each regrouped cohort had a large number of features, at least 1,409, which could cause overfitting of the model. However, after trying as many feature selection methods and machine learning models as possible, we believe that the methods finally used were relatively optimal. Finally, all CT images were acquired from one 64-slice multidetector CT scanner of the chest. In addition, most centres may no longer use 5 mm reconstruction, as chest high-resolution CT is widely considered more accurate in the assessment of pulmonary nodules. Additionally, contrast-enhanced CT is not recommended for nodule evaluation unless certain conditions apply. Therefore, these aspects may limit the generalizability of our results.
In the future, we will carry out prospective studies in a clinical setting to further validate the current results. Afterwards, the software used in our study can be optimized and installed in the work environment of physicians and radiologists to facilitate the use of proposed radiomics models in daily practice and to see if the established models could improve their diagnostic efficacy. Based on the specific design of the current study, the potential application of our results in clinical practice may be limited to patients with pulmonary nodules who may need hospitalization.
Conclusion
The current study evaluated the performance of radiomics features extracted from routine chest CT images with different reconstruction parameters in differentiating malignant and benign pulmonary nodules. The results indicated that radiomics features obtained from multiple windows demonstrated comparable performance to those from single windows. In addition, slice thickness and contrast enhancement did not substantially affect the diagnostic performance.
Data availability
Please contact author (Weimin Li) for data requests.
Abbreviations
- CT:
-
Computed tomography
- HU:
-
Hounsfield unit
- VOI:
-
Volume of interest
- SD:
-
Standard deviation
- ROC curve:
-
Receiver operating characteristic curve
- AUC:
-
Area under the curve
References
Au-Yong ITH, Hamilton W, Rawlinson J, Baldwin DR. Pulmonary nodules. BMJ. 2020;371:m3673.
Mazzone PJ, Lam L. Evaluating the patient with a pulmonary nodule.JAMA. 2022;327(3).
Anderson IJ, Davis AM. Incidental pulmonary nodules detected on CT images. JAMA. 2018;320(21):2260–1.
Callister MEJ, Baldwin DR, Akram AR, Barnard S, Cane P, Draffan J, et al. British thoracic society guidelines for the investigation and management of pulmonary nodules. Thorax. 2015;70:1–54.
Bai C, Choi CM, Chu CM, Anantham D, Chung-Man Ho J, Khan AZ, et al. Evaluation of pulmonary nodules: clinical practice Consensus Guidelines for Asia. Chest. 2016;150(4):877–93.
McWilliams A, Tammemagi MC, Mayo JR, Roberts H, Liu G, Soghrati K, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med. 2013;369(10):910–9.
Swensen SJ, Silverstein MD, Ilstrup DM, Schleck CD, Edell ES. The probability of malignancy in solitary pulmonary nodules. Application to small radiologically indeterminate nodules. Arch Intern Med. 1997;157(8):849–55.
Bera K, Braman N, Gupta A, Velcheti V, Madabhushi A. Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat Rev Clin Oncol. 2022;19(2):132–46.
Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–62.
Liu Z, Wang S, Dong D, Wei J, Fang C, Zhou X, et al. The applications of radiomics in precision diagnosis and treatment of oncology: Opportunities and challenges. Theranostics. 2019;9(5):1303–22.
Mao L, Chen H, Liang M, Li K, Gao J, Qin P, et al. Quantitative radiomic model for predicting malignancy of small solid pulmonary nodules detected by low-dose CT screening. Quant Imag Med Surg. 2019;9(2):263–72.
Xu Y, Lu L, E LN, Lian W, Yang H, Schwartz LH, et al. Application of radiomics in predicting the malignancy of pulmonary nodules in different sizes. AJR Am J Roentgenol. 2019;213(6):1213–20.
Wu W, Pierce LA, Zhang Y, Pipavath SNJ, Randolph TW, Lastwika KJ, et al. Comparison of prediction models with radiological semantic features and radiomics in lung cancer diagnosis of the pulmonary nodules: a case-control study. Eur Radiol. 2019;29(11):6100–8.
Yang X, He J, Wang J, Li W, Liu C, Gao D, et al. CT-based radiomics signature for differentiating solitary granulomatous nodules from solid lung adenocarcinoma. Lung Cancer. 2018;125:109–14.
Ligero M, Jordi-Ollero O, Bernatowicz K, Garcia-Ruiz A, Delgado-Munoz E, Leiva D, et al. Minimizing acquisition-related radiomics variability by image resampling and batch effect correction to allow for large-scale data analysis. Eur Radiol. 2021;31(3):1460–70.
Thawani R, McLane M, Beig N, Ghose S, Prasanna P, Velcheti V, et al. Radiomics and radiogenomics in lung cancer: a review for the clinician. Lung Cancer. 2018;115:34–41.
Hassani C, Varghese BA, Nieva J, Duddalwar V. Radiomics in pulmonary lesion imaging. AJR Am J Roentgenol. 2019;212(3):497–504.
Stefano A, Gioè M, Russo G, Palmucci S, Torrisi SE, Bignardi S, et al. Performance of radiomics features in the quantification of idiopathic pulmonary fibrosis from HRCT. Diagnostics. 2020;10(5):306.
Tsurugai Y, Kozuka T, Ishizuka N, Oguchi M. Relationship between the consolidation to maximum tumor diameter ratio and outcomes following stereotactic body radiotherapy for stage I non-small-cell lung cancer. Lung Cancer. 2016;92:47–52.
Lu H, Kim J, Qi J, Li Q, Liu Y, Schabath MB, et al. Multi-window CT based radiological traits for improving early detection in lung cancer screening. Cancer Manag Res. 2020;12:12225–38.
Lu H, Mu W, Balagurunathan Y, Qi J, Abdalah MA, Garcia AL, et al. Multi-window CT based radiomic signatures in differentiating indolent versus aggressive lung cancers in the National Lung Screening Trial: a retrospective study. Cancer Imaging. 2019;19(1):45.
Park S, Lee SM, Kim S, Choi S, Kim W, Do KH, et al. Performance of radiomics models for survival prediction in non-small-cell lung cancer: influence of CT slice thickness. Eur Radiol. 2021;31(5):2856–65.
He L, Huang Y, Ma Z, Liang C, Liang C, Liu Z. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep. 2016;6:34921.
Xu Y, Lu L, Sun SH, L-n E, Lian W, Yang H, et al. Effect of CT image acquisition parameters on diagnostic performance of radiomics in predicting malignancy of pulmonary nodules of different sizes. Eur Radiol. 2021;32(3):1517–27.
Acknowledgements
Not applicable.
Funding
This research is supported by Major research programs of Natural Science Foundation of China (91859203), Key R & D plan of Sichuan Provincial Department of science and technology (2021YFS0072, 2022NSFSC0785), 1.3.5 project interdisciplinary innovation project of West China Hospital of Sichuan University (ZYJC18001, ZYJC21028) and Cadre health research project of Sichuan Province (Chuanganyan 2023 − 103).
Author information
Authors and Affiliations
Contributions
WL, BC and RZ contributed to the conception and design of this study. RZ contributed to data acquisition. JS and SL contributed to data interpretation and analysis. BC and WL contributed to study supervision. RZ and JS contributed to manuscript writing and editing. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All methods were performed in accordance with the Declaration of Helsinki. The study was approved by the institutional review board of the West China Hospital of Sichuan University. Besides, informed consent was waived by the institutional review board of the West China Hospital of Sichuan University because this was a retrospective study.
Consent for publication
Not applicable.
Competing interests
The authors report no conflicts of interest in this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, R., Shi, J., Liu, S. et al. Performance of radiomics models derived from different CT reconstruction parameters for lung cancer risk prediction. BMC Pulm Med 23, 132 (2023). https://doi.org/10.1186/s12890-023-02366-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12890-023-02366-y