Abstract
Background
Synchronous multiple primary lung cancers (sMPLC) are rare forms of lung cancer, and their diagnosis remains as a significant challenge. Distinguishing sMPLC from advanced disease is important as their prognoses and therapeutic management vary dramatically.
Case presentation
The patient was a 56-year-old Chinese male who exhibited six synchronous invasive adenocarcinomas at diagnosis [T2(6)N0M0], and who achieved durable clinical benefit under adjuvant chemotherapy for 41 months following wedge resection and lobectomy. Whole-exome sequencing revealed that two lesions (L4 and L6) in the left upper lobe of the patient’s lung shared 28 nonsynonymous mutations; thus, suggesting that the lesions may have arisen from a common ancestor at the early stages of tumorigenesis, and spread into distinct histologic subtypes. Moreover, while L5 was in the same lobe as L4 and L6, it represented a distinct lineage as it did not share any mutations with other lesions. Notably, the BRAF V600E oncogenic mutation was exclusive to L5. In addition, the KRAS G12C mutation was identified in three lesions (L1-L3) located in the right lung, which may have resulted from convergent evolution.
Conclusion
We report a patient with six synchronous invasive adenocarcinomas who demonstrated durable clinical benefits under adjuvant chemotherapy following surgical treatment. While cancer staging is one of the many challenges associated with sMPLC, the data generated through next-generation sequencing can provide information on lesion origins, and thus, advance the era of precision medicine.
Similar content being viewed by others
Background
An increasing number of lung cancers (~ 15% of surgical patients) exhibit two or more malignant pulmonary lesions [1, 2]; however, synchronous multiple primary lung cancer (sMPLC) remains a rare form of lung cancer [3]. Distinguishing sMPLC from advanced disease is clinically important as the prognosis and treatment vary between the two forms of disease, and an aggressive surgical approach to sMPLC may result in rates of survival comparable to single lung cancers of similar stage [4, 5]. Thus, in the 8th edition of the Tumour, Node and Metastasis (TNM) Classification of Lung Cancer [6, 7], the International Association for the Study of Lung Cancer (IASLC) proposed that a unique staging strategy be applied to multiple ground glass opacities (GGOs) suspected of being sMPLC. While the current diagnosis of sMPLC uses the criteria defined by Martini and Melamed [8], molecular evaluation of multiple lesions has become increasingly valued for conceptually understanding the nature of such lesions, as well as the lineages (clonality) between lesions [9, 10]. The precise interpretation of the clonal origin of sMPLCs will facilitate the rationalization of treatments for sMPLC patients and improve their prognosis.
Case presentation
A 56-year-old Chinese male patient was admitted to the Hospital following the accidental discovery of GGOs in his lung during a routine physical examination. The patient was a heavy smoker for 30 years. Routine laboratory workups and the levels of serum tumor markers including carcinoembryonic antigen (CEA) were normal. Microbiological blood tests were negative, and no abnormal cells were detected during the sputum cytology test. Computed tomography (CT) scans revealed bilateral and ill-defined GGOs, including three lesions (L4-L6) in the left upper lobe (LUL), and three additional lesions, L1-L3, in the right upper lobe (RUL), right middle lobe (RML), and right lower lobe (RLL), respectively. A Corona radiata sign was observed in L1, which was in the apical segment of the RUL with the convergence of the supplying blood vessels. Cavitation was present in L3 and there was no associated lymphadenopathy in the mediastinum.
A positron emission tomography (PET) scan revealed increased 18F-fluorodeoxyglucose (FDG) avidity in these multifocal lesions. The size of the lesions ranged from 0.7 cm to 2.4 cm, with maximum standardized uptake values (SUVmax) ranging from 1.3 to 3.8. No distant metastases were detected (Fig. 1).
Chest radiograph and clinicopathologic details of the lesions. A Upper panel: Chest computed tomography scan: L1, L2 and L3 were in the right upper lobe (RUL), right middle lobe (RML) and right lower lobe (RLL), respectively. L4, L5 and L6 were all observed in the left upper lobe (LUL). No hilar or mediastinal lymphadenopathy was observed. Bottom panel: The corresponding positron emission tomography (PET-CT) scans revealed intense 18F-fluorodeoxyglucose (FDG) avidity. B Tumor lesion images. C Microscopic images of hematoxylin and eosin-stained sections representing the predominant histologic subtypes. Two representative areas are shown in (a) and (b). The corresponding histologic subtypes are described in Table 1
Each lesion was larger than 6 mm, and according to the 2017 Fleischner Society guidelines [11], all six lesions were suspected of being adenocarcinomas. Considering the National Comprehensive Cancer Network guidelines (NCCN®, 2019.V2), radical resection procedures were recommended as follows: L4-L6 subject to complete resection by a preferred anatomic left upper lobectomy, while L1-L3 were appropriate for wedge resections to preserve the patient’s lung function. Therefore, the patient underwent video-assisted thoracoscopic (VATS) right lobe wedge resections, followed by VATS LUL lobectomy 1 month later. Associated mediastinal lymph node dissection, as well as systematic lymph node sampling was also performed (thoracic lymph nodes dissected were stations 9R, 7, 10R, 11R, 2R, 4R for the right wedge resections, and stations 9 L, 7, 4 L, 5, 10 L, 11 L, 12 L for the LUL lobectomy). Pathological analyses of the surgically resected specimens revealed the six lesions as invasive pulmonary adenocarcinomas. The resection margins of the resected lung tissues were clean. L1 was reported to invade the visceral pleura (PL1), but no bronchus involvement was observed in any lesions. Additionally, no metastases were detected in any of dissected hilar or mediastinal lymph nodes.
The immunohistochemistry (IHC) data revealed that the lesions exhibited different histological subtypes, including 50% papillary + 50% acinar in L1, L3 and L5; 90% papillary in L2; 90% lepidic in L4; and 90% acinar in L6. The detailed clinicopathological characteristics of the lesions are summarized in Table 1.
According to the Martini-Melamed classification (1975), the patient was diagnosed with sMPLC [8]. The pathologic stage of L1 was T2 due to visceral pleura invasion (PL1). L3 had a maximum diameter of 24 mm and was defined as T1c, while the remaining four lesions were identified as T1b. According to the IASLC guidelines for sMPLC staging [6], the patient was determined to be pT2(6)N0M0 (T category was designated by the category of the highest T lesion, while the N and M categories were designated collectively for all lesions). Considering the high-risk factors, including visceral pleural involvement and wedge resection, and the lack of an appropriate targeted therapy, the patient received four cycles of chemotherapy with a conventional regimen of Pemetrexed (950 mg) and cisplatin (140 mg). No recurrence was observed during the 41-month follow-up period.
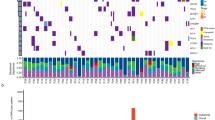
To investigate the molecular profiles of the different lesions, genomic DNA was extracted from formalin-fixed, paraffin-embedded tumor specimens using QIAamp DNA FFPE Tissue Kit (Qiagen), and library preparations were performed with KAPA Hyper Prep Kit (KAPA Biosystems). Target enrichment was performed using the xGen Exome Research Panel and Hybridization and Wash Reagents Kit (Integrated DNA Technology) according to the manufacturer’s protocol. Sequencing was performed on Illumina HiSeq4000 platform using PE150 sequencing chemistry (Illumina) [12]. To determine if lesions originated from the same ancestral clone, we required at least four nonsynonymous mutations (excluding driver mutations) to be shared by two independent tumors in a cohort of 126 lung adenocarcinomas at a probability of 0.1%, under the assumption that each patient’s tumor was of an independent origin [13]. Our data revealed that L4 and L6 shared a total of 28 nonsynonymous mutations, strongly suggesting that those lesions arose from a single clonal event during the early stages of tumorigenesis, and subsequently evolved into different histological subtypes (Fig. 2). The analysis of the alternate allelic frequency of heterozygous single nucleotide polymorphism (SNPs) further supported that the chr1q gain detected in L4 and L6 were likely of the same origin (paternal or maternal) (Figure S1).
Phylogenetic analysis of six tumor lesions. Mutational profiling using whole-exome sequencing revealed that L4 and L6 shared a total of 28 nonsynonymous mutations. L1, L2, and L3 shared three nonsynonymous mutations, including the KRAS G12C hotspot mutation. L5 represented an independent lesion of distinct lineage. The numbers of nonsynonymous mutations detected in different lesions are indicated with the representative mutations shown. Chr1q gain was detected in all lesions. The colors indicate the chr1q gain as either paternal or maternal based on the analysis of the alternate allelic frequencies of heterozygous SNPs (refer to Figure S1 for additional details)
Although L5 was in the same lobe as L4 and L6, it arose from an independent lineage, as no mutations were shared between it and the other lesions. Additionally, the BRAF V600E oncogenic mutation was exclusive to L5 (Fig. 2). Conversely, L1-L3 shared three nonsynonymous mutations, including the KRAS G12C driver mutation, which was more likely to have resulted from convergent evolution, rather than being derived from a common ancestral clone.
Discussion and conclusions
sMPLC is a rare form of lung cancer, and its diagnosis remains as a significant challenge. It is critical to distinguish sMPLC from intrapulmonary metastases as the therapeutic approach and prognosis for the two conditions are markedly different. Herein, we report a 56-year-old Chinese male patient with six synchronous invasive adenocarcinomas who achieved durable clinical benefit following adjuvant chemotherapy for 41 months after surgery. This observation was consistent with previous findings that multifocal disease is a heterogenous category where the clinical outcomes were superior to those of a single nodule at similar stages [14].
Comparisons of the molecular profiles of different lesions revealed that lesions L4 and L6, located in the LUL, likely originated from a common ancestral clone at the early stages of tumorigenesis. Despite exhibiting different histological subtypes, L4 and L6 had 28 non-silent mutations in common. Given the absence of metastatic disease in local lymph nodes, it is possible that progenitor tumor cells underwent aerogenous metastasis, which is a discontinuous spread of cancer cells from the primary tumor through the airways to adjacent or distant lung parenchyma [15]. Furthermore, although L1-L3 were in different lobes of the patient’s right lung, they shared three nonsynonymous mutations, including KRAS G12C. Given the cutoff for the number of shared mutations required to define clonality, it is less likely that L1, L2, and L3 were derived from the same ancestral clone. Rather, they may have resulted from convergent evolution.
In summary, we reported a patient with six multifocal invasive lung adenocarcinomas. Given the patient’s clinicopathological characteristics and favorable prognosis, a diagnosis of sMPLC was established. However, the presence of shared mutations between lesions suggested that some lesions may have been derived from a common ancestor at very early stages of tumorigenesis. Moreover, we showed that comprehensive genomic profiling (i.e., whole-exome sequencing) provided a new approach to understanding sMPLC in the era of precision medicine.
Availability of data and materials
All data supporting the conclusions of this case report are provided in the article.
Abbreviations
- CEA:
-
Carcinoembryonic antigen
- CT:
-
Computed tomography
- FDG:
-
18F-fluorodeoxyglucose
- GGOs:
-
Ground glass opacities
- IASLC:
-
International Association for the Study of Lung Cancer
- IHC:
-
Immunohistochemistry
- LUL:
-
Left upper lobe
- NCCN:
-
National Comprehensive Cancer Network guidelines
- RLL:
-
Right lower lobe
- RML:
-
Right middle lobe
- RUL:
-
Right upper lobe
- PET:
-
Positron emission tomography
- sMPLC:
-
synchronous multiple primary lung cancers
- SNP:
-
Single nucleotide polymorphism
- SUV:
-
Standardized uptake values
- TNM:
-
The tumour, node and metastasis classification
- VATS:
-
Video-assisted thoracoscopic
- WES:
-
Whole-exome sequencing
References
Flieder DB. Commonly encountered difficulties in pathologic staging of lung cancer. Arch Pathol Lab Med. 2007;131(7):1016–26.
Girard N, Ostrovnaya I, Lau C, Park B, Ladanyi M, Finley D, Deshpande C, Rusch V, Orlow I, Travis WD, et al. Genomic and mutational profiling to assess clonal relationships between multiple non-small cell lung cancers. Clin Cancer Res. 2009;15(16):5184–90.
Tucker MA, Murray N, Shaw EG, Ettinger DS, Mabry M, Huber MH, Feld R, Shepherd FA, Johnson DH, Grant SC, et al. Second primary cancers related to smoking and treatment of small-cell lung cancer. Lung Cancer working cadre. J Natl Cancer Inst. 1997;89(23):1782–8.
Arai J, Tsuchiya T, Oikawa M, Mochinaga K, Hayashi T, Yoshiura K, Tsukamoto K, Yamasaki N, Matsumoto K, Miyazaki T, et al. Clinical and molecular analysis of synchronous double lung cancers. Lung Cancer. 2012;77(2):281–7.
Jiang L, He J, Shi X, Shen J, Liang W, Yang C, He J. Prognosis of synchronous and metachronous multiple primary lung cancers: systematic review and meta-analysis. Lung Cancer. 2015;87(3):303–10.
Detterbeck FC, Bolejack V, Arenberg DA, Crowley J, Donington JS, Franklin WA, Girard N, Marom EM, Mazzone PJ, Nicholson AG, et al. The IASLC lung Cancer staging project: background data and proposals for the classification of lung Cancer with separate tumor nodules in the forthcoming eighth edition of the TNM classification for lung Cancer. J Thorac Oncol. 2016;11(5):681–92.
Lababede O, Meziane MA. The eighth edition of TNM staging of lung Cancer: reference chart and diagrams. Oncologist. 2018;23(7):844–8.
Martini N, Melamed MR. Multiple primary lung cancers. J Thorac Cardiovasc Surg. 1975;70(4):606–12.
Liu Y, Zhang J, Li L, Yin G, Zhang J, Zheng S, Cheung H, Wu N, Lu N, Mao X, et al. Genomic heterogeneity of multiple synchronous lung cancer. Nat Commun. 2016;7:13200.
Mansuet-Lupo A, Barritault M, Alifano M, Janet-Vendroux A, Zarmaev M, Biton J, Velut Y, Le Hay C, Cremer I, Regnard JF, et al. Proposal for a combined Histomolecular algorithm to distinguish multiple primary adenocarcinomas from intrapulmonary metastasis in patients with multiple lung tumors. J Thorac Oncol. 2019;14(5):844–56.
MacMahon H, Naidich DP, Goo JM, Lee KS, Leung ANC, Mayo JR, Mehta AC, Ohno Y, Powell CA, Prokop M, et al. Guidelines for Management of Incidental Pulmonary Nodules Detected on CT images: from the Fleischner society 2017. Radiology. 2017;284(1):228–43.
Fang W, Ma Y, Yin JC, Hong S, Zhou H, Wang A, Wang F, Bao H, Wu X, Yang Y, et al. Comprehensive Genomic Profiling Identifies Novel Genetic Predictors of Response to Anti-PD-(L)1 Therapies in Non-Small Cell Lung Cancer. Clin Cancer Res. 2019;25(16):5015–26.
Zhang XC, Wang J, Shao GG, Wang Q, Qu X, Wang B, Moy C, Fan Y, Albertyn Z, Huang X, et al. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat Commun. 2019;10(1):1772.
Trousse D, D'Journo XB, Avaro JP, Doddoli C, Giudicelli R, Fuentes PA, Thomas PA. Multifocal T4 non-small cell lung cancer: a subset with improved prognosis. Eur J Cardiothorac Surg. 2008;33(1):99–103.
Gaikwad A, Souza CA, Inacio JR, Gupta A, Sekhon HS, Seely JM, Dennie C, Gomes MM. Aerogenous metastases: a potential game changer in the diagnosis and management of primary lung adenocarcinoma. AJR Am J Roentgenol. 2014;203(6):W570–82.
Acknowledgements
We would like to thank the patient and his family for providing consent to publish. We also thank research staff and co-investigators involved in this study.
Funding
This study was supported by the Beijing Natural Science Foundation (No. 7182132), the Beijing Students’ Platform for Innovation and Entrepreneurship Training Program (No. 2019zlgc0629), and the National Key Research and Development Program of China (No. 2016YFC0901500). The funders provided financial support to this study but was not involved in in the design of the study, the collection of data, analysis, interpretation of data or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
YS and NL conceived and designed the study. ZJ, PW, and WW generated the data. YS, QO, HB, and XW analyzed the data. PL provided project administration. SZ and SL provided the resources for the study. NL supervised the study. YS, QO, MY and NL wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethic Committee of Peking Union Medical College Hospital and was conducted in accordance with the Declaration of Helsinki.
Consent for publication
Written informed consent was obtained from the patient for publication of this case report and any accompanying images. A copy of the written consent is available for review by the Editor of this journal.
Competing interests
Qiuxiang Ou, Hua Bao, Man Yu and Xue Wu are employees of Geneseeq Technology Inc., Canada. The remaining authors have no competing interests to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Heatmap of the alternate allelic frequencies of heterozygous SNPs on chromosome 1q across all lesions. The color indicates the alternate allelic frequency ranging from red to white (range: 0–100%). Lesions are clustered based on the similarities of the patterns of the alternate allelic frequency of heterozygous SNPs.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Song, Y., Jia, Z., Wu, P. et al. Comprehensive genomic profiling aids in understanding the lesion origins of a patient with six synchronous invasive lung adenocarcinomas: a case study. BMC Pulm Med 20, 80 (2020). https://doi.org/10.1186/s12890-020-1119-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12890-020-1119-9