Abstract
Background
Variation in genes implicated in homocysteine and lipid metabolism systems may influence antidepressant response for patients with major depressive disorder (MDD). This study aimed to investigate whether association of polymorphisms on the MTHFR, ApoE and ApoA4 genes with the treatment response in MDD subjects.
Methods
A total of 281 Han Chinese MDD patients received a single antidepressant drug (SSRI or SNRI) for at least 6 weeks, among whom 275 were followed up for 8 weeks. Their response to 6 weeks’ treatment and remission to 8 weeks’ treatment with antidepressant drugs was determined by changes in the 17-item Hamilton Depression Rating Scale (HARS-17) score. Single SNP and haplotype associations with treatment response were analyzed by UNPHASED 3.0.13. Logistic regression analysis was used to explore the interactions between genotypes and gender or drug type on treatment outcome, only those SNPs that had interactional association with gender or drug type were subjected to further stratified analysis.
Results
In total group, the haplotype (C-A) in MTHFR (rsl801133 and rs1801131) and the ApoE rs405509 AA genotype were significantly associated with better efficacy of antidepressants; In gender subgroups, only haplotype (C-A) in MTHFR (rsl801133 and rs1801131) was significantly associated with better efficacy of antidepressants in male subgroup; In drug type subgroup, the haplotype (C-A) in MTHFR (rsl801133 and rs1801131) and haplotype (G-C) in ApoE (rs7412 and rs405509) were associated with better efficacy of antidepressants in SNRI treated subgroup; The ApoA4 rs5092 G allele and GG genotype were associated with worse efficacy of antidepressants in SNRI treated subgroup.
Conclusions
Genetic polymorphisms in homocysteine and lipid metabolism systems are associated with antidepressant response, particularly for the interactions of the certain genetic with gender or drug type.
Similar content being viewed by others
Background
Major depressive disorder (MDD) is a common mental disorder with high rates of morbidity, recurrence, and suicide [1, 2]. Although newer antidepressant drugs are generally well tolerated and relatively effective, only 30–40% of patients achieve full remission [3]. Partial remission results in greater suffering among patients, as well as higher costs [4]. The variability in antidepressant drug response can be attributed to several factors, including genetic and environmental influences [5]. Therefore, several authors have attempted to identify variables that could predict antidepressant response, and have suggested several predictors, including clinical, psychosocial, psychophysiological, neuropsychological, neuroimaging, and genetic markers. It has also been suggested that combinations of these variables may improve predictions of treatment response [6,7,8]. Meta-analyses and consensus suggested that genetic factors are thought to play a pivotal role in individual responses to antidepressant treatment [9, 10]. Genetic research generally focuses on polymorphisms of target proteins, which are related to the mechanisms of action of antidepressant drugs.
Several studies have shown high prevalence rates of folate deficiency in depression [11, 12], presumably because of its impact on neurotransmitter synthesis, which relies on the folate-dependent one-carbon pathway. Low folate level may dampen antidepressant response, increase the risk of depressive relapse, and delay improvement in individuals treated with antidepressants [13]. Folate supplementation appears to improve the response to selective serotonin reuptake inhibitors (SSRIs) [14, 15]. One particular focus with respect to the connection between folate and depression has been the enzyme methylenetetrahydrofolate reductase (MTHFR) [16], which synthesizes 5-methyltetrahydrofolate, a carbon donor involved in the methylation of homocysteine (Hcy) to methionine. This enzyme is encoded by the MTHFR gene on chromosome 1 locus q36.3 in humans. A1298C missense mutation (cytosine-to-thymine) in the MTHFR gene results in an alanine-to valine substitution that renders MTHFR thermolabile, and may lead to elevated plasma Hcy, a vascular risk factor [17]. Many recent studies on vascular depression have suggested that chronic lesions in small blood vessels and capillaries could play a role in the pathogenesis of depression [18]. Furthermore, various lines of research have suggested MTHFR polymorphisms might enhance the environmental risks (such as low folate intake) for MDD via the interaction between genetic and environmental factors [19]. Several studies performed the association between MTHFR polymorphisms and antidepressant treatment response in different populations, which the results were contradictory [20,21,22,23,24]. Therefore, the first purpose of this study was to investigate how MTHFR polymorphisms affect antidepressant efficacy in Chinese Han MDD population.
Apolipoproteins (Apo) are lipid-binding proteins involved in the transport of lipids in plasma. Several studies suggested that changes in serum lipid composition may be related to MDD [25]. ApoE is essential for the normal catabolism of triglyceride-rich lipoprotein constituents. Accumulating evidence indicates that ApoE polymorphism affects multiple physiopathological pathways in coronary heart disease and Alzheimer’s disease (AD) [26, 27]. ApoE including epsilon 2 (ε2), ε3, and ε4 alleles, is encoded by a polymorphic gene located on chromosome 19 [28]. Although protective effects of ApoE ε2 have been reported in MDD, and ApoE ε4 may be associated with late-onset depression [29], the conclusions of previous studies were not in complete agreement. The present study focused on the ApoE gene promoter region and coding region to investigate the relationships between polymorphic loci and antidepressant efficacy. ApoA4 is another protein involved in lipid metabolic regulation, and has been shown to activate lecithin-cholesterol acyltransferase and cholesteryl esters transfer protein [30]. Data-driven analysis showed that ApoA4 has very high accuracy for discriminating individuals with remitted late-life depression (LLD) compared to never-depressed control participants [31].
A single genetic variant cannot explain the consistent variability observed in patient response to psychiatric treatment [32]. Multiple genetic variations in the Hcy and lipid metabolism pathways could explain more of the variance than a single genetic polymorphism. Therefore, in the present study, we enrolled highly homogeneous MDD patients, evaluated the efficacy of antidepressant therapy at 6 weeks response (primary endpoint) and 8 weeks remission (second endpoint) after received a single antidepressant drug, intended to confirm the association of MTHFR, ApoE and ApoA4 with MDD and antidepressant response.
Methods
Subjects
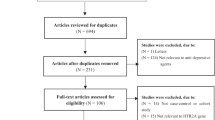
The subjects were Han Chinese inpatients from the database of Institution of Neuropsychiatry, Southeast University. Detailed information of the database referred to our previous study [33]. All patients were fulfilled the criteria for a diagnosis of MDD according to the Diagnostic and Statistical Manual of the American Psychiatric Association (DSM-IV) [34]. The inclusion and exclusion criteria (see details in SI Methods S.1) used to choose subjects can be found in our previously published study [33]. Written informed consent was obtained from all subjects or guardian participants, and the study was approved by the ethics committee of ZhongDa Hospital affiliated to Southeast University or other hospitals participating in the study. A flow chart of recruiting subjects is provided in Fig. 1.
Antidepressant treatment and clinical evaluation
According to clinical practice guidelines, a total of 281 Han Chinese patients received a single antidepressant drug (SSRI or SNRI) at least 6 weeks, among whom 275 were followed up for 8 weeks. The detail antidepressant treatment and clinical evaluation are provided in SI Methods S.2 and in our previously published study [33].
Gene selection and genotyping methods
Three candidate genes were selected based on evidence for the involvement of vascular risk factors and lipid metabolism in the mechanism of depression, including MTHFR, ApoE, and ApoA4 genes. Eight single nucleotide polymorphisms (SNPs) were detected with minor allele frequency (MAF) values of > 5% in the Asian population, according to the dbSNP and HapMap databases and using gene chips (Table S1). The genotyping methods are provided in SI Methods S.3.
Statistical analysis
Differences in clinical variables between responder and non-responder groups, as well as remission and non-remission groups, were evaluated by Student’s t test or Pearson’s χ2 test using SPSS software (version 13.0; SPSS Inc., Chicago, IL). Haploview 4.0 was used to analyze Hardy–Weinberg equilibrium (HWE), MAF, percentage of successful genotyping for each marker (%gene), and linkage disequilibrium (LD; both D’ and r2).
Population power analysis indicates that our sample of 281 subjects has 88.6% power to detect significant (p < 0.05) differences between response rates of 71 and 77%, i.e. group differences in the proportion of responders are less than 12.3%. In addition, genetic polymorphisms were associated with therapeutic effects by comparing allele, genotype and haplotype distributions of MTHFR、APOE and APOA4 between responders and non-responders, remitters and non-remitters using UNPHASED 3.0.13 (Dudbridge, 2003)). To investigate these relationships in the context of gender and drug type subgroups, we used logistic regression (SPSS 13.0) analysis to explore the interactions between genotypes and gender or drug type on treatment outcome, covariates included age, gender, drug type, and baseline 17-item Hamilton Depression Rating Scale (HDRS-17) score. Only those SNPs that had interactional association with gender or drug type were subjected to further stratified analysis. One thousand random permutations were performed using UNPHASED 3.0.13 software to correct p-values for multiple testing in the allelic, genotypic and haplotype association analyses, P < 0.05 was considered statistical significance.
Results
A total of 281 patients completed a 6-week antidepressant treatment course. Among these patients, 205 achieved a response and the response rates about 72.9%. The demographic and clinical characteristics of patients in the responder and non-responder groups are shown in Table 1. There were no significant differences between the 6-week responder and non-responder groups in gender, age, drugs used, years of education, or family history of mood disorders. However, the baseline HDRS-17 score was significantly different between these two groups (t = 2.891, P = 0.004).
A total of 275 patients completed 8-week antidepressant treatment. Among these patients, 144 achieved remission and the remission rates about 52.4%. There were no significant differences in age, number of years of education, family history, baseline HDRS-17 score, or antidepressant agents used between remission and non-remission groups (all P > 0.05), while the proportion of male patients and number of episodes were significantly higher in the non-remission group than the remission group (t = 2.381, P = 0.018 and t = − 1.983, P = 0.049, respectively), as shown in Tables 1 and 2.
Among the eight SNPs of the three genes investigated, two (ApoA4 rs5101 and rs675) were eliminated as they had MAF < 5%, while the remaining six SNPs were subjected to further statistical analyses (Table S1). LD analysis showed that two SNPs (rs1801133, rs1801131) of the MTHFR gene were in near 100% LD (D’ = 1.0, r2 = 0.177), while two other SNPs (rs405509, rs439401) of the ApoE gene were in strong LD (D’ = 0.961, r2 = 0.505). The other SNPs showed no LD.
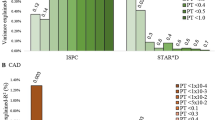
An analysis of single locus effects revealed that the ApoE rs405509 A allele and AA genotype were significantly associated with the better efficacy of antidepressants at 6 weeks in the total group (A allele: χ2 = 6.27, P = 0.012; AA genotype: χ2 = 7.41, P = 0.006), but only the AA genotype was significant after permutation testing (P* = 0.04) (Shown in Table 3). The MTHFR rs1801133 TT genotype was related to the efficacy of antidepressants, and its distribution frequency was significantly higher in the non-remission group than in the remission group (χ2 = 6.328, P = 0.012), but the result did not withstand permutation testing.
An analysis of haplotype effects demonstrated that the haplotype (C-A) in MTHFR (rs1801133 and rs1801131) was significantly associated with antidepressant response in the 8-week antidepressant in the total group (χ2 = 11.39, P = 0.0007), the result withstood permutation testing (P* = 0.02) (Shown in Table 4). The haplotypes G-A and G-C (rs7412 and rs405509), C-G (rs405509 and rs439401), and G-C-G (rs7412, rs405509, and rs439401) in ApoE were significantly associated with antidepressant response in the 6-week response (rs7412-rs405509: G-A, χ2 = 5.046, P = 0.025; G-C, χ2 = 4.313, P = 0.038; rs405509-rs439401: χ2 = 5.13, P = 0.024; rs7412-rs405509-rs439401: χ2 = 3.907, P = 0.048), but the above results did not withstand permutation testing. The non-significant results were included in supplementary materials (Table S2, S3, S4, S5).
In the logistic regression analyses of the interaction of genotype or haplotype with gender, only the haplotype of MTHFR SNPs (rs1801133 and rs1801131) showed significant result (P = 0.018, OR = 2.296), the result was shown in supplementary materials (Table S7). This haplotypes of MTHFR SNPs (rs1801133 and rs1801131) was further explored within the male/female subgroups; only the haplotype C-A was associated with treatment response in the male subgroup (χ2 = 8.767, P = 0.003), the result withstood permutation testing (P* = 0.012) (Table 4).
An exploration of the genotype or haplotype and antidepressant drug type interaction revealed that the SNP of ApoA4 rs5092, the haplotype of MTHFR SNPs (rs1801133 and rs1801131) and the haplotype of ApoE SNPs (rs7412 and rs405509) showed significant result (ApoA4 rs5092: P = 0.022, OR = 0.437; haplotype of MTHFR: P = 0.005; OR = 0.374; haplotype of ApoE: P = 0.013, OR = 0.399). The above results were shown in supplementary materials (Table S6, S7, S8). Then, the three markers were further investigated in SSRI/SNRI subgroups. As shown in Table 3, the ApoA4 rs5092 G allele and GG genotype were associated with antidepressant response, of which the antidepressant effect of G allele carriers and GG genotype were poor in SNRI subgroup (G allele: χ2 = 7.241, P = 0.007; GG genotype: χ2 = 6.964, P = 0.008), the results withstood permutation testing (G allele: P* = 0.005, GG genotype: P* = 0.019). The haplotype C-A was associated with treatment response in the SNRI subgroup (χ2 = 10.51, P = 0.001), the result withstood permutation testing (P* = 0.002) (Table 4). As shown in Table 5, the haplotype G-C in ApoE (rs7412 and rs405509) was significantly associated with antidepressant response in the SNRI subgroup (χ2 = 8.24, P = 0.0041). In comparison with the A-A haplotype, the G-C haplotype was associated with increased likelihood of a better response (OR = 1.04, 95% CI = 0.19–5.64), the result withstood permutations (P* = 0.049).
Discussion
We investigated the association of genetic variation in folate and lipid metabolism-related genes with antidepressant response in patients with MDD, and found significant effects of single polymorphisms in MTHFR, ApoE, and ApoA4.
In the MTHFR gene, C677T (rs1801133) and A1298C (rs1801131) are the most investigated SNPs associated with MDD. The C677T variant results from a single nucleotide substitution at this position, in which cytosine (C) is replaced by thymine (T) resulting a conversion of alanine to valine residue which diminishes the enzyme activity diminishes the enzyme activity. Another common polymorphism is A1298C, in which adenine (A) is replaced by cytosine (C) resulting a conversion of glutamate to alanine, which also diminishes the enzyme activity. In the present study, the haplotypes C-A of rs1801133 and rs1801131 were associated with better antidepressant effects in the whole group, and male or SNRI treated subgroups. This may have been because there are mutations in both T-A and C-C haplotypes that result in decreased MTHFR enzyme activity, increased blood Hcy levels, and decreased folate levels [35]. Bottilieri et al. reported that folic acid supplementation can protect brain function by reducing Hcy [36, 37], and also that increased levels of Hcy and/or decreased levels of folate resulted in decreased levels of S-adenosyl methionine (SAM) in cerebrospinal fluid; meanwhile, SAM as a methyl donor for serotonin (5-HT) and catecholamine pathways exerted significant antidepressant effects and was shown to have better efficacy than imipramine [38]. However, it has been suggested [39] that folic acid and vitamin B12 do not clearly enhance the efficacy of antidepressants, and that the use of folic acid and vitamin B12 can only prevent further increases in Hcy but cannot reduce its level; this may be related to differences among studies in the folic acid and vitamin B12 doses used .
This study further showed that only C-A haplotype carriers in the male but not female subgroup experienced higher antidepressant efficacy. This may have been due to the higher folate concentrations and lower Hcy levels in women [40], which compensates for reduced MTHFR enzyme activity, thus making it difficult to detect the relationship between this gene polymorphism and antidepressant efficacy. In addition, women have higher estrogen levels, and estrogen can also weaken the correlation between MTHFR polymorphism and antidepressant efficacy to some extent by lowering Hcy levels [41].
In addition, the efficacy of antidepressants was greater in C-A haplotype carriers in the SNRI subgroup. As we all known, SSRIs mainly selectively inhibit the reuptake of 5-HT by the presynaptic membrane. In contrast to SSRIs, SNRIs have a 5-HT reuptake inhibitory effect, as well as noradrenaline (NE) and mild dopamine (DA) reuptake inhibition. It has been reported that depressed patients with high plasma hcy concentrations have significantly lower concentrations of the CSF monoamine metabolites 5-hydroxyindoleacetic acid (5-HIAA), homovanillic acid (HVA), and 4-hydroxy-3-methoxyphenylethylene (MHPG) suggesting an impairment in the metabolism of 5-HT, DA and NE [42]. Therefore, SNRIs maybe have greater effects against the neurotoxicity associated with a high Hcy level. SSRIs have a single drug target, so there was little difference in efficacy between haplotypes in the SSRI subgroup.
In our present study, we also notice that single locus association analysis has found no association between SNPs rsl801131 and rsl801133 with antidepressant response, in agreement with the findings from several studies. Mei F et al. [23] found that the mutation of MTHFR gene C677T polymorphism was significantly associated with the increased risk of PSD, but not with antidepressant treatment response. Mischoulon, D et al. [24] reported that MTHFR C677T and MS A2756G polymorphism did not affect the antidepressant response of fluoxetine treatment. We speculate that a single SNP has less effect on gene function than a haplotype, which is not enough to affect the efficacy of antidepressants.
The ApoE gene plays an important role in regulating lipid metabolism and maintaining cholesterol balance. The SNP, rs7412 (C526T), is a functional site in the ApoE gene; it is mutated, resulting in replacement of arginine with cysteine. Which indicated that this polymorphism is closely related to lipid metabolism. Although rs405509 (219A/C), which is located in the promoter region upstream of the gene, has not been confirmed to be a functional site, which showed that its polymorphism is associated with antidepressant efficacy. Therefore, further studies regarding this site are required.
Studies showed that serum cholesterol levels in patients with depression were significantly lower than those in healthy people [43]. Furthermore, Sonawalla et al. reported that serum cholesterol levels have also been shown to be associated with the efficacy of antidepressants [44]. Their study found that patients with depression treated with a standard dose of fluoxetine (20 mg/d) had high serum cholesterol levels (≥ 200 mg/dl); with lower cholesterol levels, the curative effect is diminished, the tendency toward chronic disease is greater, and the possibility of recurrence is higher; related research on refractory depression reached a similar conclusion. The above studies suggested that the effects of the ApoE gene polymorphism on the efficacy of antidepressants may be related to the concentration of cholesterol in the body. Excessive cholesterol concentration may affect 5-HT transporters and/or various 5-HT receptors [45]. The function of the 5-HT neurons cytomembrane structure has an adverse effect. As another possible explanation, patients with hypercholesterolemia are more likely to have vascular and anxiety disorders, which would affect the efficacy of antidepressants [46]. However, studies have yielded inconsistent results, suggesting that the incidence of depression is lower in the higher blood lipid state, and that high blood lipids may have certain antidepressant effects. For example, Mase et al. [25] reported that low serum high-density lipoprotein (HDL-C) is a marker of suicidal behavior in depression, and may induce immune or inflammatory reactions in depression. Mischoulon et al. [47] also reported that foods rich in docosahexaenoic acid (DHA) are associated with a lower incidence of depression, while DHA deficiency (e.g., alcoholism and postpartum) is associated with a higher incidence of depression. The discrepancies between the above studies may be related to differences in the subjects and indicators of blood lipid levels.
In addition, studies on the association between the ApoE gene and depression and antidepressant efficacy have focused on the common alleles of this gene, ε2, ε3, and ε4, and some studies confirmed that the ApoE gene ε4 allele was associated with greater efficacy of antidepressants [48,49,50]. Bizzarro et al. [51] reported a significant association between AD and rs405509 CC genotypes when exploring the association between AD and an ApoE gene promoter, suggesting that rs405509 may play a role in the pathogenesis of AD. Lescai et al. [52] also reported that haplotype A-ε4, consisting of rs4905509 and ε4, can increase the risk of late-onset AD by reducing ApoE expression levels. Therefore, we hypothesized that the rs405509 polymorphism may have strong LD with other functional SNPs, such as the ε4 allele, and thus alter the biological function of the ApoE gene to influence the efficacy of antidepressants.
The function of rs5092 (29A/G) located in the promoter region of the ApoA4 gene, is still unclear and there have been few studies related to this site. However, in an exploration of gene function, the polymorphism of the ApoA1/C3/A4 gene cluster on chromosome 11q23–24 was shown to be related to blood lipids. These three genes show a high degree of identity and evolved from the same ancestral gene. The ApoA1 and ApoA4 genes have the same transcriptional direction, while ApoC3 is transcribed in the opposite direction [52]; moreover, its polymorphic variation can lead to hypertriglyceridemia [53]. This site may affect the synthesis of ApoA4, and the blood lipid level, by altering transcription of the ApoA4 gene.
Our study has limitations as follows. Firstly, there was no placebo control group in our present study, it is difficult to exclude such effects and other non-pharmaceutical factors. In further research, we can consider establishing a placebo control group to better observe the efficacy of antidepressants. Second, only three genes polymorphism in Hcy and lipid metabolic pathways were included in the present study, their treatment response may be not enough. Regarding the sites shown to be associated with the efficacy of antidepressants studies with larger sample sizes are needed to verify their accuracy and lay a foundation for personalized medicine. Finally, the reports of efficiency to treatment response due to having one of the genotypes may be affected by prior exposure to treatments for depression. Further research could enroll first episode and drug-naive depression patients to further verify the relationship between genetic polymorphisms and antidepressant drug treatment response.
Conclusions
This study demonstrates that genetic polymorphisms in homocysteine and lipid metabolism systems are associated with antidepressant response in MDD patients, particularly, with the interactions of the certain genetic with gender or drug type. The results of this study and further pharmacogenomics research may lead to individualized, more reasonable and successful new antidepressant treatment strategies.
Availability of data and materials
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study. The de-identified dataset used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- 5-HIAA:
-
5-hydroxyindoleacetic acid
- 5-HT:
-
5-hydroxytryptamine, serotonin
- AD:
-
Alzheimer’s disease
- ApoA4:
-
Apolipoprotein A4
- ApoE:
-
Apolipoprotein E
- CGI:
-
Clinical Global Impression
- DA:
-
Dopamine
- DHA:
-
Docosahexaenoic acid
- DSM-IV:
-
Diagnostic and Statistical Manual of the American Psychiatric Association
- Hcy:
-
Homocysteine
- HDL-C:
-
High-density lipoprotein
- HDRS:
-
Hamilton Depression Rating Scale
- HVA:
-
Homovanillic acid
- HWE:
-
Hardy–Weinberg equilibrium
- LLD:
-
Late-life depression
- MAF:
-
Minor allele frequency
- MDD:
-
Major depressive disorder
- MHPG:
-
4-hydroxy-3-methoxyphenylglycol
- MTHFR:
-
Methylenetetrahydrofolate reductas
- NE:
-
Norepinephrine
- SAM:
-
S-adenosyl methionine
- SNPs:
-
Single nucleotide polymorphisms
- SNRI:
-
Serotonin norepinephrine reuptake inhibitor
- SSRIs:
-
Serotonin reuptake inhibitors
- TESS:
-
Treatment Emergent Symptom Scale
References
Vandeleur CL, Fassassi S, Castelao E, Glaus J, Strippoli MF, Lasserre AM, Rudaz D, Gebreab S, Pistis G, Aubry JM, et al. Prevalence and correlates of DSM-5 major depressive and related disorders in the community. Psychiatry Res. 2017;250:50–8.
Huang Y, Wang Y, Wang H, Liu Z, Yu X, Yan J, Yu Y, Kou C, Xu X, Lu J, et al. Prevalence of mental disorders in China: a cross-sectional epidemiological study. Lancet Psychiatry. 2019;6(3):211–24.
Kelley ME, Dunlop BW, Nemeroff CB, Lori A, Carrillo-Roa T, Binder EB, Kutner MH, Rivera VA, Craighead WE, Mayberg HS. Response rate profiles for major depressive disorder: characterizing early response and longitudinal nonresponse. Depress Anxiety. 2018;35(10):992–1000.
Tranter R, O'Donovan C, Chandarana P, Kennedy S. Prevalence and outcome of partial remissionin depression. J Psychiatry Neurosci. 2002;27(4):241–7.
Lohoff FW, Ferraro TN. Pharmacogenetic considerations in the treatment of psychiatric disorders. Expert Opin Pharmacother. 2010;11(3):423–39.
Mocking RJ, Figueroa CA, Rive MM, Geugies H, Servaas MN, Assies J, Koeter MW, Vaz FM, Wichers M, van Straalen JP, et al. Vulnerability for new episodes in recurrent major depressive disorder: protocol for the longitudinal DELTA-neuroimaging cohort study. BMJ Open. 2016;6(3):e009510.
Bigdeli TB, Ripke S, Peterson RE, Trzaskowski M, Bacanu SA, Abdellaoui A, Andlauer TF, Beekman AT, Berger K, Blackwood DH, et al. Genetic effects influencing risk for major depressive disorder in China and Europe. Transl Psychiatry. 2017;7(3):e1074.
Vall E, Wade TD. Predictors of treatment outcome in individuals with eating disorders: a systematic review and meta-analysis. Int J Eating Disord. 2015;48(7):946–71.
Fabbri C, Hosak L, Mössner R, Giegling I, Mandelli L, Bellivier F, Claes S, Collier DA, Corrales A, Delisi LE, et al. Consensus paper of the WFSBP Task Force on Genetics: Genetics, epigenetics and gene expression markers of major depressive disorder and antidepressant response. World J Biol Psychiatry. 2017;18(1):5-28.
Kato M, Serretti A. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol Psychiatry. 2010;15(5):473–500.
Alpert JE, Mischoulon D, Nierenberg AA, Fava M. Nutrition and depression: focus on folate. Nutrition. 2000;16(7–8):544.
Tiemeier H, van Tuijl HR, Hofman A, Meijer J, Kiliaan AJ, Breteler MM. Vitamin B12, folate, and homocysteine in depression: the Rotterdam study. Am J Psychiatr. 2002;159(12):2099.
Jain R, Jackson WC. Beyond the resistance: how novel neurobiological understandings of depression may lead to advanced treatment strategies. J Clin Psychiatry. 2012;73(11):e30.
Alpert JE, Mischoulon D, Rubenstein GEF, Bottonari K, Nierenberg AA, Fava M. Folinic acid (Leucovorin) as an adjunctive treatment for SSRI-refractory depression. Ann Clin Psychiatry. 2002;14(1):33–8.
Hintikka J, Tolmunen T, Tanskanen A, Viinamaki H. High vitamin B12 level and good treatment outcome may be associated in major depressive disorder. BMC Psychiatry. 2003;3:17.
McGuffin P, Knight J, Breen G, Brewster S, Boyd PR, Craddock N, Gill M, Korszun A, Maier W, Middleton L, et al. Whole genome linkage scan of recurrent depressive disorder from the depression network study. Hum Mol Genet. 2005;14(22):3337–45.
Cortese C, Motti C. MTHFR gene polymorphism, homocysteine and cardiovascular disease. Public Health Nutr. 2001;4(2B):493–7.
Santos M, Kovari E, Hof PR, Gold G, Bouras C, Giannakopoulos P. The impact of vascular burden on late-life depression. Brain Res Rev. 2009;62(1):19–32.
Wan L, Li Y, Zhang Z, Sun Z, He Y, Li R. Methylenetetrahydrofolate reductase and psychiatric diseases. Transl Psychiatry. 2018;8(1):242.
Shen X, Wu Y, Guan T, Wang X, Qian M, Lin M, Shen Z, Sun J, Zhong H, Yang J, et al. Association analysis of COMT/MTHFR polymorphisms and major depressive disorder in Chinese Han population. J Affect Disord. 2014;161:73–8.
Jamerson BD, Payne ME, Garrett ME, Ashley-Koch AE, Speer MC, Steffens DC. Folate metabolism genes, dietary folate and response to antidepressant medications in late-life depression. Int J Geriatr Psychiatry. 2013;28(9):925–32.
Lanctôt KL, Rapoport MJ, Chan F, Rajaram RD, Strauss J, Sicard T, McCullagh S, Feinstein A, Kiss A, Kennedy JL, et al. Genetic predictors of response to treatment with citalopram in depression secondary to traumatic brain injury. Brain Inj. 2010;24(7–8):959–69.
Mei F, Wu Y, Ding G, Pan F, Chen L, Wu J. Association of methylenetetrahydrofolate reductase gene 677C>T polymorphism with post-stroke depression risk and antidepressant treatment response in Han Chinese. JPMA. 2018;68(7):888–92.
Mischoulon D, Lamon-Fava S, Selhub J, Katz J, Papakostas GI, Iosifescu DV, Yeung AS, Dording CM, Farabaugh AH, Clain AJ, et al. Prevalence of MTHFR C677T and MS A2756G polymorphisms in major depressive disorder, and their impact on response to fluoxetine treatment. CNS Spectrums. 2012;17(2):76–86.
Maes M, Smith R, Christophe A, Vandoolaeghe E, Gastel AV, Neels H, Demedts P, Wauters A, Meltzer HY. Lower serum high-density lipoprotein cholesterol (HDL-C) in major depression and in depressed men with serious suicidal attempts: relationship with immune-inflammatory markers. Acta Psychiatr Scand. 1997;95(3):212.
Nikolac Perkovic M, Pivac N. Genetic markers of Alzheimer's disease. Adv Exp Med Biol. 2019;1192:27–52.
Chen W, Jin F, Cao G, Mei R, Wang Y, Long P, Wang X, Ge W. ApoE4 may be a promising target for treatment of coronary heart disease and Alzheimer's disease. Curr Drug Targets. 2018;19(9):1038–44.
Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proc Natl Acad Sci U S A. 2006;103(15):5644–51.
F.Sharpley C: The role of genes (and environmental stress) in depression: an update. Curr Psychiatr Rev 2011, 7(2):84–95.
Kim YS, Gu BH, Choi BC, Kim MS, Song S, Yun JH, Chung MK, Choi CH, Baek KH. Apolipoprotein A-IV as a novel gene associated with polycystic ovary syndrome. Int J Mol Med. 2013;31(3):707–16.
Diniz BS, Lin CW, Sibille E, Tseng G, Lotrich F, Aizenstein HJ, Reynolds CF, Butters MA. Circulating biosignatures of late-life depression (LLD): towards a comprehensive, data-driven approach to understanding LLD pathophysiology. J Psychiatr Res. 2016;82:1–7.
Pickar D, Rubinow K. Pharmacogenomics of psychiatric disorders. Trends Pharmacol Sci. 2001;22(2):75–83.
Xu Z, Zhang Z, Shi Y, Pu M, Yuan Y, Zhang X, Li L, Reynolds GP. Influence and interaction of genetic polymorphisms in the serotonin system and life stress on antidepressant drug response. J Psychopharmacol. 2012;26(3):349–59.
Association AP. Diagnostic and statistic manual of mental disorders 5 (DSM-5). Diabetes. 1994;11(1):97–8.
Kamphuis MH, Geerlings MI, Grobbee DE, Kromhout D. Dietary intake of B (6-9-12) vitamins, serum homocysteine levels and their association with depressive symptoms: the Zutphen elderly study. Eur J Clin Nutr. 2008;62(8):939–45.
Bottiglieri T. Homocysteine and folate metabolism in depression. Prog Neuro-Psychopharmacol Biol Psychiatry. 2005;29(7):1103–12.
Herrmann W, Obeid R. Biomarkers of folate and vitamin B (12) status in cerebrospinal fluid. Clin Chem Lab Med. 2007;45(12):1614–20.
Bell KM, Plon L, Bunney WE Jr, Potkin SG. S-adenosylmethionine treatment of depression: a controlled clinical trial. Am J Psychiatry. 1988;145(9):1110–4.
Christensen H, Aiken A, Batterham PJ, Walker J, Mackinnon AJ, Fenech M, Hickie IB. No clear potentiation of antidepressant medication effects by folic acid+vitamin B12 in a large community sample. J Affect Disord. 2011;130(1–2):37–45.
Nanri A, Mizoue T, Matsushita Y, Sasaki S, Ohta M, Sato M, Mishima N. Serum folate and homocysteine and depressive symptoms among Japanese men and women. Eur J Clin Nutr. 2010;64(3):289–96.
Kalra DK. Homocysteine and cardiovascular disease. Curr Atheroscler Rep. 2004;6(2):101–6.
Bottiglieri T, Laundy M, Crellin R, Toone BK, Carney MW, Reynolds EH. Homocysteine, folate, methylation, and monoamine metabolism in depression. J Neurol Neurosurg Psychiatry. 2000;69(2):228–32.
Kim EJ, Hong J, Hwang J-W. The association between depressive mood and cholesterol levels in Korean adolescents. Psychiatry Investig. 2019;16(10):737–44.
Sonawalla SB, Papakostas GI, Petersen TJ, Yeung AS, Smith MM, Sickinger AH, Gordon J, Israel JA, Tedlow JR, Lamon-Fava S, et al. Elevated cholesterol levels associated with nonresponse to fluoxetine treatment in major depressive disorder. Psychosomatics. 2002;43(4):310–6.
Pucadyil TJ, Chattopadhyay A. Cholesterol modulates the antagonist-binding function of hippocampal serotonin1A receptors. Biochim Biophys Acta Biomembr. 2005;1714(1):35–42.
Suárez Bagnasco M. Psychological issues and cognitive impairment in adults with familial hypercholesterolemia. Fam Pract. 2017;34(5):520–4.
Mischoulon D, Fava M. Docosahexanoic acid and omega-3 fatty acids in depression. Psychiatric Clin North Am. 2000;23(4):785–94.
Murphy GM, Kremer C, Rodrigues H, Schatzberg AF. The apolipoprotein E epsilon4 allele and antidepressant efficacy in cognitively intact elderly depressed patients. Biol Psychiatry. 2003;54(7):665–73.
Schmand B, Hooijer C, Jonker C, Lindeboom J, Havekes LM. Apolipoprotein E phenotype is not related to late-life depression in a population-based sample. Soc Psychiatry Psychiatr Epidemiol. 1998;33(1):21–6.
Mauricio M, O'Hara R, Yesavage JA, Friedman L, Kraemer HC, Van De Water M, Murphy GM Jr. A longitudinal study of apolipoprotein-E genotype and depressive symptoms in community-dwelling older adults. Am J Geriatr Psychiatry. 2000;8(3):196–200.
Bizzarro A, Seripa D, Acciarri A, Matera MG, Pilotto A, Tiziano FD, Brahe C, Masullo C. The complex interaction between APOE promoter and AD: an Italian case-control study. Eur J Hum Genet. 2009;17(7):938–45.
Lescai F, Chiamenti AM, Codemo A, Pirazzini C, D'Agostino G, Ruaro C, Ghidoni R, Benussi L, Galimberti D, Esposito F, et al. An APOE haplotype associated with decreased epsilon4 expression increases the risk of late onset Alzheimer's disease. J Alzheimer’s Dis. 2011;24(2):235–45.
Groenendijk M, Cantor RM, de Bruin TW, Dallinga-Thie GM. The apoAI-CIII-AIV gene cluster. Atherosclerosis. 2001;157(1):1–11.
Acknowledgements
We are sincerely grateful to all the subjects who participated in the study.
Funding
The study has been partly supported by the National Key Research and Development Program of China (No. 2016YFC1306700, Zhijun Zhang), the National Natural Science Key Foundation of China (No. 81830040, Zhijun Zhang), Jiangsu Provincial Medical Outstanding Talent (No. JCRCA2016006, Zhijun Zhang) and Scientific Research Program of Jiangsu Provincial Commission of Health and Family Planning (No. H2017007, Baoyu Yuan). The funding body did not contribute to the design of the study or the collection, analysis, or interpretation of data, or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
ZZ and YY designed the research protocol, SX, XZ and PM analyzed data; YB and ZZ wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The authors state that the study was established according to the ethical guidelines of the Helsinki Declaration and was approved by the ethics committee of ZhongDa Hospital affiliated to Southeast University or other hospitals participating in the study. Written informed consent was obtained from all subjects or guardian participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that Yonggui Yuan as a member of Editorial Board.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yuan, B., Sun, X., Xu, Z. et al. Influence of genetic polymorphisms in homocysteine and lipid metabolism systems on antidepressant drug response. BMC Psychiatry 20, 408 (2020). https://doi.org/10.1186/s12888-020-02798-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12888-020-02798-4