Abstract
Background
Subchromosomal deletions and duplications are the leading cause of congenital malformations and mental retardation in children. With the recent clinical application of genomic microarrays in the evaluation of patients with developmental delays and congenital malformations, it has led to the discovery of several new microdeletion and microduplication syndromes. However, there are no published reports involving patients with both microduplications in the 9p21.1-p24.3 region and microdeletions in the 7p22.1-p22.3 region.
Case presentation
We report an infant with an autosomal abnormality confirmed by conventional karyotype combined with copy number variations sequencing (CNV-seq), showing the patient with an unbalanced translocation. The karyotype of the patient was 46, XX, der (7)t (7;9) (p22; p21) and CNV-seq results showed an approximately 32.34-Mb duplication in 9p21.1-p24.3 (200000-32540000) and an approximately 3.3-Mb deletion in 7p22.2-p22.3 (40000-3340000).
Conclusions
The patient carried an unbalanced translocation 46, XX, der (7)t (7;9) (p22; p21) derived from her mother. The clinical presentation is closely related to the size and position of the missing and duplicated chromosomes. To our knowledge, the simultaneous occurrence of de novo partial trisomy 9p(9p21.1-p24.3) and partial monosomy 7p (7p22.2-p22.3) has not previously been reported up until now. The present study additionally demonstrated that CNV-seq combined with karyotype is able to reliably detect unbalanced submicroscopic chromosomal aberrations.
Similar content being viewed by others
Background
Trisomy 9p syndrome is a rare disorder, first reported by Rethoré et al. [1] in 1970. Most reported cases of trisomy 9p are accompanied by partial deletions of other chromosomes. It is characterized by multi-organ system involvement, including craniofacial anomalies, cardiac, genitourinary, skeletal and central nervous system (CNS) abnormalities [2]. Karyotype analysis is the “gold standard” for diagnosing chromosomal aberrations. It usually detects abnormal chromosome numbers and structural abnormalities such as deletions, duplications, translocations and inversions of large segments of 5–10 Mb or more, but not deletions and duplications of small chromosomal segments [3, 4]. With the development of molecular genetic techniques, the CNV-seq technique can detect micro-repeats and micro-deletions as small as tens of kilobases, and can determine the size of duplicated or missing fragments and their location on chromosomes, which is a powerful complement to the traditional karyotype analysis. In this study, we combined karyotype analysis of chromosome G and CNV-seq to perform cytogenetic and molecular genetic tests in a patient with growth retardation and mental retardation with congenital multiple malformations. in order to identify the origin of chromosomal abnormalities and analyze the relationship between chromosomal structural abnormalities and clinical phenotypes, thus providing a strong basis for clinical diagnosis and genetic counseling.
Case presentation
The proband was a 4-month-old female born to a 29-year-old father and a 27-year-old mother via vaginal delivery at 38 gestational weeks. During pregnancy, no specific problems were identified. The patient was hospitalized in a local hospital for 7 days after birth for “respiratory distress syndrome”. She was found to have slow weight gain since birth, with a birth weight of 2.5 kg and a current weight of 3.5 kg, accompanied by poor feeding, dry vomiting, minor crying, and bruised lips while crying. In order to seek further medical treatment, the patient was admitted to Children’s Hospital Affiliated to Zhengzhou University (Zhengzhou, China) at 4-months-old for “malnutrition”. Since the onset of the disease, the patient had poor mental response, poor appetite, and slightly dilute stool. The examination showed a stunted development, malnutrition, and thin subcutaneous fat. There were peculiar facial features including wide eye spacing, small jaw, high palatal arch, left eyelid ptosis, hawkish nose and low ear position. The left thumb was attached on top of the palm, and the little fingers on both hands were flexed and deformed. The breath sounds of both lungs were coarse and a grade 3/6 murmur could be heard in the precordial region. Congenital heart disease was suspected. This diagnosis was followed by transthoracic atrial septal defect closure and arterial catheterization for treatment.
Peripheral blood samples were obtained from the patient, her sister and the parents for examination of chromosomes by metaphase G-banding and CNV-seq. The Children’s Hospital Affiliated to Zhengzhou University Ethics Committee approved the sample collection procedures and the family gave written informed consent. Chromosome karyotype analyses under sterile conditions was conducted on cultured lymphocytes according to standard protocols. Colchicine was added after 72 h of culture and cells were harvested after 1 h, after which they were filmed, G-banded for color development, photographed with a GSL120 fully automated scanner. 20 cells were counted and 5 karyotypes were analyzed by applying karyotype analysis software. Chromosomal karyotypes were determined according to the International System of Human Cytogenetics Nomenclature ISCN (2020). CNV-seq assays standard procedures were used isolate the genomic DNA of the proband and the parents from whole blood using PerkinElmer Chemagic 360 fully automated nucleic acid extractor. The library was constructed using the “Rapid PCR-free library construction technology” (Berrygenomics, Inc., Beijing, China). Detection of copy number variations (CNVs) was conducted by NextSeq CN500 (Illumina, Inc., USA) high-throughput sequencer. The sequencing type was single-end 36-base sequencing. The measured sequence fragments were compared to the known human reference genome (hg19). Analysis was performed using Konoan data analysis software (Berrygenomics Genetic Diagnostics, Inc., Hangzhou, China), and CNVs were detected with a resolution of 100 kb or more. The copy number variations were compared with the Database of Genomic Variants (DGV), the Database of Genomic Variantion and Phenotype in Humans using Ensembl Resources (DECIPHER), Online Mendelian Inheritance in Man (OMIM), and The Clinical Genome Resource (ClinGen) to annotate the reported disease-causing genes by comparison and analysis. According to the American College of Medical Genetics and Genomics (ACMG) guidelines and the CNVs diagnostic guidelines, CNVs are rated as 5 levels of risk: pathogenic, possibly pathogenic, benign, probably benign, and of unknown significance [5, 6].
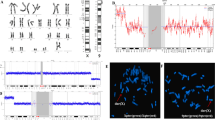
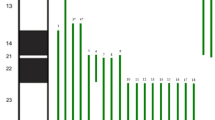
The karyotype of the patient was 46, XX, der (7) t (7;9) (p22; p21) mat (Fig. 1A). The karyotype of the patient’s mother indicated a balanced translocation karyotype: 46, XX, t (7;9) (p22; p21) (Fig. 1B). Her sister and father exhibited a normal karyotype. The CNV-seq analysis revealed a 32.34 Mb duplication in the 9p21.1p24.3 (200000-32540000) (hg19) region, involving 100 OMIM genes, and a 3.30 Mb deletion in the 7p22.2p22.3 (40000-3340000) (hg19) region, involving 30 OMIM genes (Fig. 2A and B). By searching databases such as Decipher, OMIM, DGV, and ClinGen, clinical phenotype matching and interpretation of genetic patterns were performed for cases that had been reported in the databases. The results showed that the two CNVS of the child were reported in various databases and involved many genes. Finally, according to the ACMG guidelines and the guidelines for the diagnosis of CNVs, they were both classified as pathogenic CNVs.
The results of karyotype analysis of chromosomes. (A)Karyotype of the patient. The karyotype of the patient indicated an abnormal karyotype: 46, XX, der (7)t(7;9) (p22; p21) mat. The arrow indicates the derived chromosome 7. (B)Karyotype of the mother of the patient. The karyotype of the mother of the patient indicated an abnormal karyotype: 46, XX, der (7)t(7;9) (p22; p21). The arrows indicated chromosomes with balanced translocation
Discussion and conclusions
The present case report demonstrated that the patient carried an unbalanced translocation inherited from the mother who was a balanced translocation carrier, which resulted in partial trisomy for 9p (spanning ~ 32.34 Mb) and partial monosomy for 7p (spanning ~ 3.30 Mb). To the best of our knowledge, the present study is the first report of an unbalanced translocation involving chromosomes 7p and 9p.
9p trisomy is often caused by heterozygous segregation of familial chromosomal translocations. Most reports include deletions of other chromosomes. Common phenotypes of trisomy 9p include growth and language intellectual disability, abnormal ear position, hypertelorism, bulbous nose, low mouth angle, and abnormal hand and foot finger development[2]. The severity of the partial trisomy 9p phenotype was correlated with the length of the repeat in the short arm of chromosome 9 and the repeat region. Duplications in the 9p13-p21 region have less effect on mental development whereas some genes associated with mental development (DOCK8, FOXD4, VLDLR, etc.) are present in the 9p22-9p24 region where the very low density lipoprotein receptor gene (VLDLR) transduces a variety of extracellular signals across the neural cell membrane into the CNS, regulates synaptic plasticity and is important for specific learning and memory functions in the hippocampus [7]. The duplications in p21.1-p24.3 of chromosome 9 in the patient involved“trisomy 9 syndrome, which contains 100 OMIM genes. There are multiple patients in the Decipher database carrying pathogenic or potentially pathogenic variants that partially overlap with this CNV interval. It has been reported in the literature that the main clinical manifestations of patients with 9p22-p24 duplication include short stature, microcephaly, peculiar facial features, and congenital heart disease [8,9,10]. The clinical phenotype of this patient was generally consistent with the phenotypes reported in the literature, with the addition of other typical phenotypic features, such as a hawkish nose and presence of unilocular ptosis. This discrepancy may be due to the fact that it carries a 9p repeat fragment that is less consistent with the above literature. Another possibility may be due to its coexistence with a heterozygous deletion of approximately 3.30 Mb in the 7p22.2p22.3 region, which contains 30 OMIM genes including 11 morbidity-associated genes (BRAT1, FAM20C, EIP3B, LFNG, INTS1, etc.). INTS1 and BRAT1 genes are located at 7p22.3 and are associated with uniform neurodevelopmental disorders [11]. It has been reported in the literature [12] that the main clinical phenotype of patients with 7p22.2p22.3 deletion is a peculiar facial appearance, with developmental delay in speech, etc. Both chromosomal copy number variants in this patient have been reported frequently in patient databases, with the involvement of additional genes, and all were determined to be pathogenic CNVs according to ACMG guidelines. The reported phenotypes correlate with the patient’s phenotype, but the reported phenotypes were more variable and our patient was comparatively younger. Many phenotypes require further clinical excavation, verification and follow-up observations. Whether haploinsufficiency of any OMIM gene necessarily leads to a clinical phenotype requires further summary and follow-up of additional cases. In this case, the patient had both trisomy 9p and monosomy 7p. It is possible that abnormal alterations in these two chromosomes interact to form a specific phenotype.
Phenotypic outcomes such as recurrent spontaneous abortion, embryonic arrest and multiple neonatal malformations tend to manifest in carriers of chromosomal balanced translocation. The results of karyotype analysis suggested that the mother of the patient was a 46, XX, t (7; 9) (p22; p21) balanced translocation carrier, which was the direct cause of the microdeletion of segment 7p22.2p22.3 and the duplication of segment 9p21.1p24.3 in the patient. The reason for this is that the probability for a balanced translocation carrier to produce normal gametes is extremely low, with a theoretical probability of obtaining phenotypically normal offspring of only 1/9 [13, 14] and an actual probability of about 1/3. The mechanism of CNV formation in this patient may be due to the instability of the parental translocation chromosome break sites. For such children, prenatal diagnosis and preimplantation genetic diagnosis (PGD) are the main ways to reduce birth defects, so that children with genetic defects and various congenital anomalies can be detected early, and intrauterine treatment can be performed at the right time for those who are eligible and can be corrected, and those who cannot be corrected can have their pregnancies terminated in time to reduce the birth of defective children. In addition, prenatal diagnosis allows the chromosomes of both parents to be known in order to obtain the karyotype, breakpoints, and mode of inheritance of the translocation, providing a basis for genetic counseling and prenatal diagnosis for the incidence of translocation chromosome carriers and pregnancy outcome as well as revealing the genetic etiology of the clinical manifestations of the affected children. During the genetic counseling process, a comprehensive analysis should be performed in conjunction with several factors such as the type of translocation in the translocation carrier, the chromosomes involved in the translocation, and the location of the breakpoint of the translocation.
In conclusion, the occurrence of concurrent partial trisomy 9q (9p21.1p24.3) and partial monosomy 7p (7p22.2p22.3) has not previously been reported up to now. This study combined the application of karyotype analysis and CNV-seq to finally confirm the diagnosis for the patient. The use of CNV-seq and karyotype may facilitate a sensitive and powerful approach towards the diagnosis of submicroscopic unbalanced genomic rearrangements. This study clarified the origin and formation mechanism of CNV in children, and analyzed the relationship between chromosomal structure abnormalities and patient phenotype. Due to the clear mechanism of its occurrence and high risk of recurrence, clinical genetic counseling was presented to the patient’s mother where she was advised to undergo prenatal examination and diagnosis in the event of future pregnancies.
Data availability
The datasets that support the conclusions of this article are available by request to the corresponding author. We do not make participants’ data publicly available due to data protection restrictions and participant confidentiality.
Abbreviations
- CNV-seq:
-
Copy number variants sequencing
- CNS:
-
central nervous system
- DGV:
-
Database of Genomic Variants
- DECIPHER:
-
Database of Genomic Variantion and Phenotype in Humans using Ensembl Resources
- OMIM:
-
Online Mendelian Inheritance in Man
- ClinGen:
-
Clinical Genome Resource
- ACMG:
-
American College of Medical Genetics and Genomics
References
Rethoré MO, Larget-Piet L, Abonyi D, Boeswillwald M, Berger R, Carpentier S, et al. 4 cases of trisomy for the short arm of chromosome 9. Individualization of a new morbid entity. Ann Genet. 1970;13(4):217–32.
Li M, Glass J, Du X, Dubbs H, Harr MH, Falk M, et al. Trisomy 9 mosaic syndrome: sixteen additional patients with new and/or less commonly reported features, literature review, and suggested clinical guidelines. Am J Med Genet A. 2021;185(8):2374–83.
Qian YQ, Fu XY, Wang XQ, Luo YQ, Chen M, Yan K, et al. feasible Diagn approach translocation carrier indication Prod conception Mol Cytogenet. 2018;11:12.
Qian YQ, Wang XQ, Chen M, Luo YQ, Yan K, Yang YM, et al. Detection of fetal subchromosomal aberration with cell-free DNA screening led to diagnosis of parental translocation: review of 11344 consecutive cases in a university hospital. Eur J Med Genet. 2019;62(2):115–23.
Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13(7):680–5.
South ST, Lee C, Lamb AN, Higgins AW, Kearney HM. ACMG Standards and Guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications: revision 2013. Genet Med. 2013;15(11):901–9.
Qiu S, Korwek KM, Weeber EJ. A fresh look at an ancient receptor family: emerging roles for low density lipoprotein receptors in synaptic plasticity and memory formation. Neurobiol Learn Mem. 2006;85(1):16–29.
Guilherme RS, Meloni VA, Perez AB, Pilla AL, de Ramos MA, Dantas AG, et al. Duplication 9p and their implication to phenotype. BMC Med Genet. 2014;15:142.
Guanciali Franchi P, Calabrese G, Morizio E, Modestini E, Stuppia L, Mingarelli R et al. FISH analysis in detecting 9p duplication (p22p24) Am J Med Genet, 2000. 90(1): p. 35 – 7.
Fujimoto A, Lin MS, Schwartz S. Direct duplication of 9p22–>p24 in a child with duplication 9p syndrome. Am J Med Genet. 1998;77(4):268–71.
Xie HH, Liu T, Zhang JB, Zhai JF, Liu Y. Partial trisomy 16q and partial monosomy 7p of a fetus derivated from paternal balanced translocation: a case report. Med (Baltim). 2021;100(7):e24382.
Yu AC, Zambrano RM, Cristian I, Price S, Bernhard B, Zucker M et al. Variable developmental delays and characteristic facial features-A novel 7p22.3p22.2 microdeletion syndrome? Am J Med Genet A, 2017. 173(6): p. 1593–1600.
Scarinci R, Anichini C, Vivarelli R, Berardi R, Pucci L, Rosaia L, et al. [Correlation of the clinical phenotype with a pericentric inversion of chromosome 9]. Boll Soc Ital Biol Sper. 1992;68(3):175–81.
Allderdice PW, Kaita H, Lewis M, McAlpine PJ, Wong P, Anderson J, et al. Segregation of marker loci in families with an inherited paracentric insertion of chromosome 9. Am J Hum Genet. 1986;39(5):612–7.
Acknowledgements
The authors wish to thank the patient’s family and all the colleagues who contributed to the management of the family.
Funding
This study was funded by Henan Province Medical Science and Technology Tackling Program Joint Co-Construction Project (LHGJ20220757).
Author information
Authors and Affiliations
Contributions
RL and LFL conceptualized the main structure of the report, and drafted, reviewed and revised the manuscript. CJW and ZHZ prepared the clinical trial data. DXL contributed in performing genetic analysis. ZJX and DZ reviewed and revised the manuscript. All the authors read and approved the final manuscript as submitted.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study has been approved by the Ethics Committee of the Children’s Hospital of Zhengzhou University. The patient’s parents had provided written informed consent to participate in genetic analysis. We confirm that all methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Written informed consent for publication of identifiable information/ images in open access journal was obtained from the parents of the participant.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, R., Wang, C., Zhang, Z. et al. Partial trisomy 9p and partial monosomy 7p of an infant inherited from maternal balanced translocation: a case report. BMC Pediatr 23, 168 (2023). https://doi.org/10.1186/s12887-023-03986-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12887-023-03986-3