Abstract
Purpose
To describe the genetic and clinical features of nineteen patients from eleven unrelated Chinese pedigrees with OPA1-related autosomal dominant optic atrophy (ADOA) and define the phenotype-genotype correlations.
Methods
Detailed ophthalmic examinations were performed. Targeted next-generation sequencing (NGS) was conducted in the eleven probands using a custom designed panel PS400. Sanger sequencing and cosegregation were used to verify the identified variants. The pathogenicity of gene variants was evaluated according to American College of Medical Genetics and Genomics (ACMG) guidelines.
Results
Nineteen patients from the eleven unrelated Chinese ADOA pedigrees had impaired vision and optic disc pallor. Optical coherence tomography showed significant thinning of the retinal nerve fiber layer. The visual field showed varying degrees of central or paracentral scotoma. The onset of symptoms occurred between 3 and 24 years of age (median age 6 years). Eleven variants in OPA1 were identified in the cohort, and nine novel variants were identified. Among the novel variants, two splicing variants c.984 + 1_984 + 2delGT, c.1194 + 2 T > C, two stop-gain variants c.1937C > G, c.2830G > T, and one frameshift variant c.2787_2794del8, were determined to be pathogenic based on ACMG. A novel splicing variant c.1316-10 T > G was determined to be likely pathogenic. In addition, a novel missense c.1283A > C (p.N428T) and two novel splicing variants c.2496G > A and c.1065 + 5G > C were of uncertain significance.
Conclusions
Six novel pathogenic variants were identified. The findings will facilitate genetic counselling by expanding the pathogenic mutation spectrum of OPA1.
Similar content being viewed by others
Introduction
Autosomal dominant optic atrophy (ADOA, MIM#165,500) is one of the most common hereditary optic neuropathies, with an estimated prevalence ranging from 1:10,000 to 1:30,000 worldwide [1,2,3,4]. It is mainly characterized by progressive symmetric painless visual impairment and optic atrophy caused by the degeneration of retinal ganglion cells and their axons in early childhood. More than 20% of the ADOA patients may present with one or more additional features, such as neurosensory hearing loss, progressive external ophthalmoplegia, ptosis, peripheral neuropathy, cataracts or ataxia, syndromic Parkinsonism and dementia[5], which are named ADOA ‘plus’[6]. The penetrance of ADOA varies from 43 to 100% in different families with different mutations [3].
To date, 13 genes, including OPA1, OPA2, OPA3, OPA4, DNM1L, OPA6, TMEM126A, OPA8, ACO2, RTN4IP1, YME1L1, AFG3L2 [7], SSBP1 [8] have been found to be associated with hereditary optic atrophy. The OPA1 gene variants contribute 57 ~ 89% to ADOA [9, 10]. Localized on 3q28-q29, the gene OPA1 spans more than 100 kb and includes 31 exons, namely exons 1 to 29, exon 4b, and exon 5b. Among them, exon 29 is nonprotein-coding. Alternative splicing of exons 4, 4b and 5b generates eight transcript isoforms [11], which are widely present in tissues, while the expression levels in different tissues vary [12]. The main isoform expressed in the human retina is isoform 1 (NM_015560.2) without 4b and 5b [9], and it was originally identified and frequently used to describe most variants in the OPA1 gene.
The OPA1 gene encodes a mitochondrial dynamin-related GTPase family protein [13] of 907 ~ 1015 amino acids, located in the inner mitochondrial membrane and involved in the formation of mitochondrial cristae and mitochondrial fusion. The OPA1 protein contains five domains, three of which are conserved domains: the GTPase domain (exons 8–15), dynamin central region (exons 16–24), and GTPase effector domain (exons 27–28) [14].
Having enabled an efficient and credible detection of gene mutations [15], high-throughput sequencing (HTS) makes the extensive molecular diagnosis possible. Here, we aimed to describe the genetic and clinical features of nineteen patients with identified OPA1 variants in eleven unrelated Chinese ADOA pedigrees and determine the pathogenicity of these novel variants.
Methods
Participants & clinics
This research was conducted in accordance with the tenets of the Declaration of Helsinki and approved by the Ethics Committee of the Henan Eye Hospital. All the participants or their guardians signed informed consents forms (HNEECKY-2019 (15), 15 October 2019). The eleven unrelated Chinese families with optic atrophy were outpatients of the Henan Eye Hospital. Detailed clinical data, including age of onset, disease duration, family history, best-corrected visual acuity (BCVA), fundus photography, optical coherence tomography (SS-OCT, VG200, Henan China), visual field (VF), full-field electroretinogram (ERG) and visual evoked potential (VEP), were collected.
DNA sample collection & targeted next-generation sequencing
Peripheral blood samples were collected from the participants of the eleven ADOA families and preserved at -20 ℃ before further analysis. Total genomic DNA was extracted with a whole blood DNA extraction kit (TIANGEN, Beijing, China) and quantified with Qubit 4.0.
Targeted next-generation sequencing was performed with a custom designed panel PS400 [16,17,18], which contains 376 inherited retinal dystrophies and other posterior segment eye disease genes, the 50 bp next to the exons and the known pathogenic/likely pathogenic variants in the introns of the genes. Genomic DNA was randomly sonicated into fragments of approximately 150–200 bp to prepare Illumina paired-end libraries. The DNA fragments were end-repaired, and an extra ‘A’ base was added to the 3' end. Illumina adapters were ligated to the ends of the DNA fragments, and PCR amplification was performed for each sample. PCR amplification was used to enrich the target gene with the specific index and RNA probe. The DNA libraries were quantified by Qubit 4.0. The enriched DNA libraries were sequenced on an Illumina Nextseq500 system (Illumina, San Diego, CA).
Bioinformatics analysis and sanger sequencing verification
The raw reads were aligned to the human genome reference (USUC hg19) using the Burrows Wheeler Aligner (BWA). Single-nucleotide variants (SNVs) and InDels (Insertions and Deletions) were called by Atlas-SNP2 and Atlas-Indel, respectively. Variant-filtering was based on public and in-house SNP databases, including the Exome Aggregation Consortium database (ExAC), the Genome Aggregation Database (gnomAD, http://gnomad.broadinstitute.org/), Human Genetic Variation Database (HGVD, http://www.genome.med.kyoto-u.ac.jp/SnpDB/), the 1000 Genomes Project database (1000 Genomes, http://browser.1000genomes.org), and the UK10K databases, as well as our internal database, with allele frequency cut-offs of 2% and 0.1% for recessive and dominant variants, respectively. We used three commercial software programs, XYGeneRanger 2.0 (Xunyin, Shanghai, China), TGex (LifeMap Sciences, Alameda, CA, USA) and Efficient Genosome Intepration System, EGIS (SierraVast Bio-Medical Technology Co., Ltd, Shanghai, China), to analyse the pathogenicity of the mutations. The nonsynonymous and splicing variants were analysed by in silico gene function prediction software such as PolyPhen-2 (Polymorphism Phenotyping v2, http://genetics.bwh.harvard.edu/pph2), SIFT (Sorting Intolerant From Tolerant, http://sift.jcvi.org) [19], MutationTaster (http://www.mutationtaster.org) [20], PROVEAN (Protein Variation Effect Analyzer, http://provean.jcvi.org/index.php), and CADD (Combined Annotation Dependent Depletion, https://cadd.gs.washington.edu/) [21]. NetGene2 (http://www.cbs.dtu.dk/services/NetGene2), NNSplice (http://www.fruitfly.org/seq_tools/splice.html) and FSPLICE (http://www.softberry.com/berry.phtml?topic=fsplice&group=programs&subgroup=gfind) were used to predict the splicing defects. Genomic Evolutionary Rate Profiling (GERP)[22] software was applied to identify constrained loci. Project HOPE (http://www.cmbi.umcn.nl/hope) was used to analyze the structural effects of missense variants. The variants were further validated and segregated by Sanger sequencing from all available family members. The pathogenicities of all variants were classified according to the American College of Medical Genetics and Genomics (ACMG)[23] standards and guidelines.
Results
Clinical characteristics of the ADOA patients
There were 33 participants in the 11 pedigrees, and 21 carried variants in OPA1. Nineteen of them, including 9 males and 10 females, presented the phenotype. The clinical characteristics of the patients are summarized in Table 1. The age of the patients ranged from 4 to 64 years, with a median age of 12 years. The onset of symptoms occurred between 3 and 24 years of age, and the median was 6 years. Painless progressively symmetric insidious vision loss was the chief complaint in all 19 patients. The disease onset of most cases was from childhood, but the reported age of onset was 24 years for patient F7- III:3. F9-IV:2 showed dyserythrochloropsia. For the most recent evaluation, BCVA varied from finger count to 0.5, and optic atrophy with temporal pallor or diffuse pallor was seen in most cases. Figure 1 shows the clinical features of a representative DOA patient F6-II:2. Two heterozygous mutation carriers, F2-I:1 and F8-I:1, however, showed a normal phenotype with neither vision loss nor optic atrophy. OCT was performed on the father F2 I-1 but not F8 I-1, so a subclinical optic nerve atrophy can’t be excluded for F8 I-1. OCT showed significant thinning of the retinal nerve fiber layer (RNFL). VF showed varying degrees of predominantly central or paracentral scotoma, even temporal hemianopsia, for patient F11-II:2, who presented with external ophthalmoplegia in addition to optic atrophy. No other extraocular neurologic features were observed.
Genetic analysis of the ADOA patients
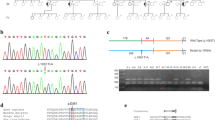
We performed targeted next-generation sequencing (NGS) in the 11 ADOA families, and identified five splicing variants, c.2496G > A, c.984 + 1_984 + 2delGT, c.1065 + 5G > C, c.1194 + 2 T > C and c.1316-10 T > G, two frameshift deletion variants, c.2708_2711delTTAG and c.2787_2794del8, three stop-gain variants, c.2830G > T, c.1937C > G and c.112C > T, and a missense variant, c.1283A > C (p.N428T) in the OPA1 gene. Figure 2 shows the pedigrees and sequencing results of the 11 ADOA families. The disease showed autosomal-dominant inheritance, and 7 of the variants were significantly cosegregation with the disease. F2-II:2 and her father carried the same heterozygous c.2708_2711delTTAG frameshift variant. F8-II:1 and his father shared the same c.1937 C > G (p.S646X) stop-gain variant. However, F2-II:2 and F8-II:1 showed typical optic nerve atrophy, while their fathers had a normal phenotype. F4-III:1 carried a de novo heterozygous splicing variant c.984 + 1_984 + 2delGT. F11-II:2 carried a de novo heterozygous splicing variant c.1316-10 T > G. Neither of their parents had the same variants, but neither of their paternities was checked.
Pedigrees and sequencing results of the 11 OPA1-related ADOA families. A F1-I:1, F1-II:3 and F1-III:3 carried the heterozygous c.2787_2794del8 variant. B F2-I:1 and F2-II:2 carried the heterozygous c.2708_2711delTTAG variant. C F3-II:2 and F3-III:3 carried the heterozygous c.2496G > A variant. D F4-III:1 carried the de novo heterozygous c.984 + 1_984 + 2delGT variant. E F5-III:1 and F5-IV:1 carried the heterozygous c.1283A > C (p.N428T) variant. F F6-II:2 carried the heterozygous c.2830G > T variant. G F7-III:3 carried the heterozygous c.1065 + 5G > C variant. H F8-I:1 and F8-II:1 carried the heterozygous c.1937C > G variant. I F9-III:3 and F9-IV:2 carried the heterozygous c.1194 + 2 T > C variant. J F10-III:1, F10-III:3 and F10-IV:1 carried the heterozygous c.112C > T variant. K F11-II:2 and F11-III:1 carried the heterozygous c.1316-10 T > G variant
All 11 OPA1 mutations identified are demonstrated in the schematic diagram of the OPA1 gene (Ref. NM_015560.2) and protein (Fig. 3). Among these variants, five (5/11, 45.5%) are located at the GTPase domain, three (3/11, 27.3%) at the GTPase effector (GE) domain, two (2/11, 18.2%) at the dynamin central region domain and one (1/11, 9.1%) at the basic domain. In addition, six of these mutations occurred in the exons, while the other occurred in introns.
All 11 OPA1 mutations identified in this study are shown in the schematic diagram of the OPA1 gene (Ref. NM_015560.2, below) and OPA1 protein (above). Missense, splicing, stop-gain and frameshift deletion mutations are coloured in black, blue, red and yellow, respectively. CC, coiled coil domain; GE, GTPase effector domain
Pathogenicity analyses of OPA1 variants in the 11 Chinese patients are summarized in the Table 2. According to ACMG standards and guidelines, of the 11 OPA1 variants, nine novel undocumented variants were identified. Five novel variants were classified as pathogenic: two splicing variants, c.984 + 1_984 + 2delGT and c.1194 + 2 T > C, two stop-gain variants, c.1937C > G and c.2830G > T, and one frameshift variant, c.2787_2794del8. A novel splicing variant c.1316-10 T > G was determined to be likely pathogenic. Among the above, the two variants c.984 + 1_984 + 2delGT and c.1316-10 T > G were de novo. A novel missense variant, c.1283A > C (p.N428T), and two novel splicing variants, c.2496G > A and c.1065 + 5G > C, were of uncertain significance. In addition, the pathogenic stop-gain variant c.112C > T (OPA1_000236, Variant #0000760813 (NC_000003.11: g.193332591C > T, OPA1(NM_015560.2):c.112C > T)—Global Variome shared LOVD) has been reported in ADOA families [24].
Discussion
ADOA represents autosomal dominant optic atrophy, and 57 ~ 89% of ADOA cases are caused by variants in the OPA1 gene. According to the Leiden Open Variation Database (LOVD, https://www.lovd.nl/), the original eOPA1 database, more than 400 OPA1 pathogenic variants have been reported (https://databases.lovd.nl/shared/genes/OPA1). Among them, 28% are missense variants, 24% are associated with altered splicing, 22% are frameshift variants, 15% are nonsense variations, and 7% are deletions [25]. In this study, 11 probands from 11 unrelated Chinese ADOA families presented varying vision defects and optic disc pallor, and were all identified as carrying heterozygous OPA1 variants. Not consistent with LOVD, among these variants, splicing variants (5/11, 45.5%) were the most common mutation type, followed by the stop-gain (3/11, 27.3%), frameshift deletion (2/11, 18.2%), and missense variants (1/11, 9.1%), which were the most common in the LOVD OPA1 database. The difference may be related to race or the smaller sample size in this study. On the other hand, missense mutations are more likely to develop ADOA ‘plus’ phenotypes [26, 27], while the only missense variant in our study did not present with plus phenotypes other than ocular signs.
The penetrance of ADOA varies from 43 to 100% in different families with different mutations [3]. We also observed the incomplete penetrance of ADOA. The heterozygous mutation carriers F2-I:1 (c.2708_2711delTTAG) and F8-I:1 (c.1937C > G) had a normal phenotype with neither decreased visual acuity nor optic atrophy. Therefore, for those who have atypical phenotypes, genetic tests are essential to make a correct diagnosis.
Haploinsufficiency and dominant negative effects contribute to the pathogenesis of the OPA1-related ADOA [28]. Haploinsufficiency indicates that pathogenic OPA1 variants lead to impaired OPA1 functions by reducing the expression of OPA1 protein. In our study, regardless of where the variant loci and protein domains were located, all splicing, stop-gain and frameshift deletion variants, were predicted to cause reduced OPA1 protein. Haploinsufficiency should be their pathogenesis. In addition, the variant types seemed to have no correlation with the severity of vision defects, which was in agreement with Xu et al. [29].
On the other hand, previous studies have shown that some missense mutations in the GTPase domain of OPA1 do not reduce the expression of OPA1. The mutated protein may compete with the wild-type protein and inhibit the function of OPA1, resulting in a dominant negative effect, thereby interfering with OPA1 functions. F5-IV:1, a 10-year-old boy, and his father F5-III:1 both carried the heterozygous c.1283A > C (p.N428T) missense variant and presented with vision impairment and optic atrophy. Just as most missense pathogenic variants reported for the OPA1 gene were clustered in the highly conserved GTPase domain, the only missense variant c.1283A > C (p.N428T) in the current study was also located in exon 13 in the GTPase domain, which may be caused by a dominant negative effect.
Clearly, the documentation of dominant negative effect diseases is of great significance. It has been confirmed in animal studies and clinical trials that autosomal recessive inherited diseases could benefit from gene augmentation therapy. However, gene therapy of autosomal dominant diseases remains a challenge. To date, three Opa1 mouse models carrying the truncation mutations (c.1051C > T, c.1065 + 5G > A, c.2708-2711delTTAG) [30,31,32,33] and showing haploinsufficiency have been tested for gene therapy. Unlike haploinsufficiency, simple augmentation of normal OPA1 levels may not be effective for gene therapy of missense mutations because of a dominant-negative mode of action [34]. Therefore, it is necessary to develop mouse models carrying missense mutations in OPA1 and presenting dominant negative effects. In addition, missense mutations tend to develop ADOA “plus” [27], which also impairs multiple systems such as the musculoskeletal system, nervous system and circulation system. It is also a challenge to develop therapeutic strategies for diseases affecting multiple organs.
The GTPase domain, dynamin central region, and GTPase effector domain are conserved [14]. More than 50% of the pathogenic OPA1 variants are located in the GTPase domain and the GTPase effector domain (exons 27–28) [35]. Similar to the database, among the variants identified in this study, which mainly affected the coding sequence and exon–intron boundaries of the gene (six of these mutations occurred at the exons, while the other occurred at the introns), eight (8/11, 72.7%) were located in the two domains, highlighting the importance of these domains in OPA1 protein functions. Impairment of GTPase activity could decrease the stability of the inner mitochondrial membrane structure and membrane potential because of proton leakage [36]. Additionally, according to a meta-analysis of genotype–phenotype analysis of OPA1-related ADOA, the most common exon involved was exon 27 [37]. Two frameshift variants, c.2787_2794del8 and c.2708_2711delTTAG, were in exon 27, which might lead to the premature termination of OPA1 protein synthesis and truncated proteins and protein degradation or nonsense-mediated mRNA decay. It is worth mentioning that the hotspot variant OPA1 gene c.2708_2711delTTAG identified in this study has been reported multiple times and could account for approximately 10% of ADOA [9, 38, 39]. In addition, variants in the CC domain were rarely reported in the LOVD database, and there was no variant identified in the CC domain in this study due to the small sample size.
According to ACMG standards and guidelines, we defined eight pathogenic variants (one variant was classified as likely pathogenic) of the 11 OPA1 variants. Three novel variants, including a missense variant c.1283A > C (p.N428T), and two splicing variants, c.2496G > A and c.1065 + 5G > C, were of uncertain significance. The HOPE online software revealed that the missense variant c.1283A > C (p.N428T) could change the physico-chemical parameters or structure of the OPA1 protein. Alavi et al. reported a mutation c.1065 + 5G > A, which is in the same location as c.1065 + 5G > C. They confirmed in a mouse model carrying c.1065 + 5G > A in the Opa1 gene that c.1065 + 5G > A induced a skipping of exon 10 during transcript processing and led to an in-frame deletion of 27 amino acid residues in the GTPase domain [30]. Multiple software programs (NetGene2, NNSplice and FSPLICE) predicted that the splicing variant c.2496G > A changed the donor splice sites.
In conclusion, we identified nine novel and two reported variants of the OPA1 gene from 11 unrelated Chinese ADOA families. All 19 patients had varying impaired vision and signs. In addition, we defined six novel pathogenic variants of the 11 OPA1 variants. Medical genetic tests are essential to make a diagnosis for those who have atypical phenotypes because of incomplete penetrance. An integrated comprehension of the clinical and genetic spectrum of ADOA would certainly advance therapeutic approaches.
Availability of data and materials
Data presented in this study are contained within the article.
References
Eiberg H, Kjer B, Kjer P, Rosenberg T. Dominant optic atrophy (OPA1) mapped to chromosome 3q region I Linkage analysis. Hum Mol Genet. 1994;3(6):977–80.
Kjer B, Eiberg H, Kjer P, Rosenberg T. Dominant optic atrophy mapped to chromosome 3q region. II. Clinical and epidemiological aspects. Acta Ophthalmol Scand. 1996;74(1):3–7.
Yu-Wai-Man P, Griffiths PG, Burke A, Sellar PW, Clarke MP, Gnanaraj L, Ah-Kine D, Hudson G, Czermin B, Taylor RW, et al. The prevalence and natural history of dominant optic atrophy due to OPA1 mutations. Ophthalmol. 2010;117(8):1538–46 1546 e1531.
Fraser JA, Biousse V, Newman NJ. The neuro-ophthalmology of mitochondrial disease. Surv Ophthalmol. 2010;55(4):299–334.
Carelli V, Musumeci O, Caporali L, Zanna C, La Morgia C, Del Dotto V, Porcelli AM, Rugolo M, Valentino ML, Iommarini L, et al. Syndromic parkinsonism and dementia associated with OPA1 missense mutations. Ann Neurol. 2015;78(1):21–38.
Yu-Wai-Man P, Griffiths PG, Gorman GS, Lourenco CM, Wright AF, Auer-Grumbach M, Toscano A, Musumeci O, Valentino ML, Caporali L, et al. Multi-system neurological disease is common in patients with OPA1 mutations. Brain. 2010;133(Pt 3):771–86.
Caporali L, Magri S, Legati A, Del Dotto V, Tagliavini F, Balistreri F, Nasca A, La Morgia C, Carbonelli M, Valentino ML, et al. ATPase Domain AFG3L2 Mutations Alter OPA1 Processing and Cause Optic Neuropathy. Ann Neurol. 2020;88(1):18–32.
Jurkute N, Leu C, Pogoda HM, Arno G, Robson AG, Nurnberg G, Altmuller J, Thiele H, Motameny S, Toliat MR, et al. SSBP1 mutations in dominant optic atrophy with variable retinal degeneration. Ann Neurol. 2019;86(3):368–83.
Delettre C, Griffoin JM, Kaplan J, Dollfus H, Lorenz B, Faivre L, Lenaers G, Belenguer P, Hamel CP. Mutation spectrum and splicing variants in the OPA1 gene. Hum Genet. 2001;109(6):584–91.
Toomes C, Marchbank NJ, Mackey DA, Craig JE, Newbury-Ecob RA, Bennett CP, Vize CJ, Desai SP, Black GC, Patel N, et al. Spectrum, frequency and penetrance of OPA1 mutations in dominant optic atrophy. Hum Mol Genet. 2001;10(13):1369–78.
Del Dotto V, Fogazza M, Carelli V, Rugolo M, Zanna C. Eight human OPA1 isoforms, long and short: What are they for? Biochim Biophys Acta Bioenerg. 2018;1859(4):263–9.
Olichon A, Elachouri G, Baricault L, Delettre C, Belenguer P, Lenaers G. OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted function in apoptosis. Cell Death Differ. 2007;14(4):682–92.
Lenaers G, Neutzner A, Le Dantec Y, Juschke C, Xiao T, Decembrini S, Swirski S, Kieninger S, Agca C, Kim US, et al. Dominant optic atrophy: Culprit mitochondria in the optic nerve. Prog Retin Eye Res. 2021;83:100935.
Belenguer P, Pellegrini L. The dynamin GTPase OPA1: more than mitochondria? Biochim Biophys Acta. 2013;1833(1):176–83.
Glockle N, Kohl S, Mohr J, Scheurenbrand T, Sprecher A, Weisschuh N, Bernd A, Rudolph G, Schubach M, Poloschek C, et al. Panel-based next generation sequencing as a reliable and efficient technique to detect mutations in unselected patients with retinal dystrophies. Europ J Hum Genet. 2014;22(1):99–104.
Fu L, Li Y, Yao S, Guo Q, You Y, Zhu X, Lei B. Autosomal Recessive Rod-Cone Dystrophy Associated With Compound Heterozygous Variants in ARL3 Gene. Front Cell Dev Biol. 2021;9:635424.
Zhu Q, Rui X, Li Y, You Y, Sheng XL, Lei B. Identification of Four Novel Variants and Determination of Genotype-Phenotype Correlations for ABCA4 Variants Associated With Inherited Retinal Degenerations. Front Cell Dev Biol. 2021;9:634843.
Zhang L, Li Y, Qin L, Wu Y, Lei B. Autosomal Recessive Retinitis Pigmentosa Associated with Three Novel REEP6 Variants in Chinese Population. Genes (Basel). 2021;12(4):537.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81.
Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–6.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–5.
Cooper GM, Stone EA, Asimenos G, Program NCS, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15(7):901–13.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24.
Nakamura M, Lin J, Ueno S, Asaoka R, Hirai T, Hotta Y, Miyake Y, Terasaki H. Novel mutations in the OPA1 gene and associated clinical features in Japanese patients with optic atrophy. Ophthalmol. 2006;113(3):483-488.e481.
Le Roux B, Lenaers G, Zanlonghi X, Amati-Bonneau P, Chabrun F, Foulonneau T, Caignard A, Leruez S, Gohier P, Procaccio V, et al. OPA1: 516 unique variants and 831 patients registered in an updated centralized Variome database. Orphanet J Rare Dis. 2019;14(1):214.
Amati-Bonneau P, Valentino ML, Reynier P, Gallardo ME, Bornstein B, Boissiere A, Campos Y, Rivera H, de la Aleja JG, Carroccia R, et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy “plus” phenotypes. Brain. 2008;131(Pt 2):338–51.
Amati-Bonneau P, Milea D, Bonneau D, Chevrollier A, Ferre M, Guillet V, Gueguen N, Loiseau D, de Crescenzo MA, Verny C, et al. OPA1-associated disorders: phenotypes and pathophysiology. Int J Biochem Cell Biol. 2009;41(10):1855–65.
Bonifert T, Karle KN, Tonagel F, Batra M, Wilhelm C, Theurer Y, Schoenfeld C, Kluba T, Kamenisch Y, Carelli V, et al. Pure and syndromic optic atrophy explained by deep intronic OPA1 mutations and an intralocus modifier. Brain. 2014;137(Pt 8):2164–77.
Xu X, Wang P, Jia X, Sun W, Li S, Xiao X, Hejtmancik JF, Zhang Q. Pathogenicity evaluation and the genotype-phenotype analysis of OPA1 variants. Mol Genet Genomics. 2021;296(4):845–62.
Alavi MV, Bette S, Schimpf S, Schuettauf F, Schraermeyer U, Wehrl HF, Ruttiger L, Beck SC, Tonagel F, Pichler BJ, et al. A splice site mutation in the murine Opa1 gene features pathology of autosomal dominant optic atrophy. Brain. 2007;130(Pt 4):1029–42.
Davies VJ, Hollins AJ, Piechota MJ, Yip W, Davies JR, White KE, Nicols PP, Boulton ME, Votruba M. Opa1 deficiency in a mouse model of autosomal dominant optic atrophy impairs mitochondrial morphology, optic nerve structure and visual function. Hum Mol Genet. 2007;16(11):1307–18.
Sarzi E, Angebault C, Seveno M, Gueguen N, Chaix B, Bielicki G, Boddaert N, Mausset-Bonnefont AL, Cazevieille C, Rigau V, et al. The human OPA1delTTAG mutation induces premature age-related systemic neurodegeneration in mouse. Brain. 2012;135(Pt 12):3599–613.
Sarzi E, Seveno M, Piro-Megy C, Elziere L, Quiles M, Pequignot M, Muller A, Hamel CP, Lenaers G, Delettre C. OPA1 gene therapy prevents retinal ganglion cell loss in a Dominant Optic Atrophy mouse model. Sci Rep. 2018;8(1):2468.
Del Dotto V, Fogazza M, Lenaers G, Rugolo M, Carelli V, Zanna C. OPA1: How much do we know to approach therapy? Pharmacol Res. 2018;131:199–210.
Neuhann T, Rautenstrauss B. Genetic and phenotypic variability of optic neuropathies. Expert Rev Neurother. 2013;13(4):357–67.
Olichon A, Landes T, Arnaune-Pelloquin L, Emorine LJ, Mils V, Guichet A, Delettre C, Hamel C, Amati-Bonneau P, Bonneau D, et al. Effects of OPA1 mutations on mitochondrial morphology and apoptosis: relevance to ADOA pathogenesis. J Cell Physiol. 2007;211(2):423–30.
Ham M, Han J, Osann K, Smith M, Kimonis V. Meta-analysis of genotype-phenotype analysis of OPA1 mutations in autosomal dominant optic atrophy. Mitochondrion. 2019;46:262–9.
Chen J, Xu K, Zhang X, Jiang F, Liu L, Dong B, Ren Y, Li Y. Mutation screening of mitochondrial DNA as well as OPA1 and OPA3 in a Chinese cohort with suspected hereditary optic atrophy. Invest Ophthalmol Vis Sci. 2014;55(10):6987–95.
Chen Y, Jia X, Wang P, Xiao X, Li S, Guo X, Zhang Q. Mutation survey of the optic atrophy 1 gene in 193 Chinese families with suspected hereditary optic neuropathy. Mol Vis. 2013;19:292–302.
Acknowledgements
The author would like to thank all participants and their families for participating in this study and their agreement to use their clinical data in this study.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81770949 and 82071008) and Henan Key Laboratory of Ophthalmology and Vision Science.
Author information
Authors and Affiliations
Contributions
Conceptualization, B.L.; methodology, J.H. and Y.L.; software, J.H., Y.L.; validation, B.L. and Y.L.; formal analysis, J.H. and Y.L.; investigation, J.H.; resources, B.L. and K.F.; data curation, J.H. and Y.L.; writing—original draft preparation, J.H.; writing—review and editing, B.L.; visualization, J.H.; supervision, B.L.; project administration, B.L.; funding acquisition, B.L. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Subjects older than 16 gave their informed consent for inclusion before they participated in the study. For children aged between 4 and 16, informed consent from their parent or legal guardian was obtained. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Henan Eye Hospital (HNEECKY-2019 (15), 15 October 2019).
Consent for publication
Written informed consent was obtained from the patients (or their parents or legal guardian in the case of children under 16) to publish this paper.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Han, J., Li, Y., You, Y. et al. Autosomal dominant optic atrophy caused by six novel pathogenic OPA1 variants and genotype–phenotype correlation analysis. BMC Ophthalmol 22, 322 (2022). https://doi.org/10.1186/s12886-022-02546-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12886-022-02546-0