Abstract
Background
We present 3 members of a family with macular dystrophy, originally diagnosed as Stargardt disease, with a significantly variable age at onset, caused by a heterozygous mutation in CRX.
Case presentation
A 43-year-old female with bull’s eye maculopathy, whose sister was diagnosed with Stargardt disease previously at another centre, was found to have a single ABCA4 variant. Further examination of the family revealed that the asymptomatic father was also affected, indicating a dominant pattern of inheritance. In addition, the ABCA4 variant was not identified in the sister originally diagnosed with Stargardt disease. Next generation sequencing identified a heterozygous c.121C > T, p.R41W missense mutation in CRX in all 3 affected members.
Conclusions
We describe a common phenotype, but with variable age at onset, with autosomal dominant inheritance and reduced penetrance in a family found to have a pathogenic sequence variant in CRX. This illustrates the importance of panel based molecular genetic testing accompanied by family studies to establish a definitive diagnosis.
Similar content being viewed by others
Background
Macular dystrophies (MD) may have certain recognisable clinical features that can pinpoint a specific diagnosis enabling a secure phenotype classification. However, in some cases patients present with retinal features that are shared by different conditions, known as phenocopies. In addition, since the advent of NGS, more than one potential causative gene variant may be identified in a family with an inherited retinal dystrophy thus making a definitive diagnosis difficult. However molecular genetic testing, accompanied by careful family studies with segregation of the variant with disease, can enable a conclusive diagnosis. This is important for prognosis, genetic counselling, and potential therapeutic interventions such as gene therapy.
Case presentation
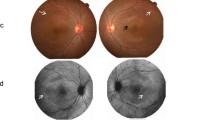
A 43-year-old white female (II:2) presented with reduced vision in her right eye, inadvertently noticed when closing her left eye. On examination visual acuities were 6/60 OD and 6/7.5 OS. Fundoscopy revealed bilateral central atrophy, seen clearly on autofluorescence imaging (AF) (Fig. 1a and b). Eight years later AF imaging showed a preserved small central spot of AF, surrounded by a reduced AF signal corresponding to the area of atrophy, which increased in size over time, with a ring of AF. (Fig. 1c-f). Optical coherence tomography (OCT) imaging, 10 years after initial presentation, showed loss of the ellipsoid zone (EZ) (Fig. 1g and h). These features were consistent with a macular dystrophy, with features of bull’s eye maculopathy (BEM). Electrophysiology testing showed that the pattern electro-retinogram was significantly affected in both eyes; the rod electro-retinogram (ERG) showed slight increase in implicit times of both ‘a’ and ‘b’ waves with significant reduction of the ‘b’ wave amplitude. The photopic and 30 Hz flicker stimuli were normal; the EOG light rise was borderline reduced in the right eye, and normal in the left (Fig. 2). In view of the family history of Stargardt macular dystrophy (STGD1) and the BEM phenotype genetic testing for ABCA4 variants was undertaken. Sanger sequencing was used to screen all 50 exons and approximately 50 base pairs of flanking intronic regions (standard practice at the time) in the proband. This identified a single previously reported pathogenic variant c.4685 T > C, p.(I1562T) [1], in the proband (II/2, Fig. 3), but a second variant was not identified; (at that time, 15–30% of STGD patients were reported as having only one detectable mutation in ABCA4 [2,3,4]). A family survey was then performed including her sister (II:4) and father (I:2), who were consented and examined. Her sister (II:4), previously diagnosed with STGD1 aged 18 years old, was re-examined at 46 years. Visual acuities were 6/60 right and left, and fundoscopy showed marked central atrophy (Fig. 4 a and b). The atrophy seen on AF and OCT imaging were similar to that seen in the proband, but were more advanced (Fig. 4c to f). Multimodal imaging at follow up 4 years later, aged 54 years, showed a further increase in central atrophy (Fig. 4g-l). Their father (I:2), aged 70 years old, was asymptomatic with visual acuities of 6/6 right, and left with a normal appearing fundus (Fig. 5a and b). However, his AF imaging showed abnormalities of central stippled reduction in signal, consistent with early macular atrophy (Fig. 5 c and d), but less advanced than that seen in his daughters (Figs. 1 and 4). Nine years after his diagnosis, aged 79, his visual acuities had deteriorated to 6/60 right, and left and he had developed central atrophic changes similar to his daughters (Fig. 5e-j). Electrophysiology results were not available for II:4, or I:II. None of the family had any co-existing systemic disorders.
Images of proband at different time points: a and b show AF at age of 51, 8 years after first presentation with a central preserved spot of AF, surrounded by atrophy, and a ring of AF external to the atrophy. c and d show wide field Optos images at age of 59 (16 years after presentation) with central atrophy; e and f show AF at the same time showing enlargement of the decreased AF signal; g and h OCT showing loss of the EZ between the yellow arrows
Key: Autofluorescence AF, Optical coherence tomography (OCT), Ellipsoid Zone (EZ).
Electrophysiology testing results for the proband (a) Pattern ERG with the reduction in amplitude in both eyes (b) Scotopic ERG showing increased ‘a’ and ‘b’ wave implicit times, and reduction in ‘b” wave amplitude in both eyes (c) Photopic ERG showing normal ‘a’ and ‘b’ wave latencies and normal ‘a’ and ‘b’ wave amplitudes for both flash and 30 Hz flicker stimuli (d) electro-oculogram showing borderline Arden Index in the right eye and normal in the left eye
Imaging of the proband’s younger sister (II/4). Images (a to f) at review aged 56, at 38 years after diagnosis. Optos imaging showing atrophic maculae (a and b). Autofluorescence (AF) shows loss of signal centrally corresponding to atrophy with a surrounding band of increased AF within the arcades (c and d. optical coherence tomography (OCT) imaging showing the temporal extent of the loss of the ellipsoid zone demarcated by yellow arrows. e and f. Images (g to l) showing increased atrophy after a 4-year interval. Optos imaging shows increased bilateral macular atrophy (g and h). AF imaging shows extension of patchy AF surrounding the central atrophy with a small preserved foveal remnant (i and j). OCT showing further extension of the loss of the ellipsoid zone demarcated by the yellow arrows
Imaging of proband’s father (I/2) Images (a to d) at age 73 (asymptomatic. Colour images of both maculae which appeared normal (a and b). Autofluorescence (AF) revealed central patchy reduction in AF signal, consistent with early atrophy (c and d). Images E to J of I:2 9 years later. Optos imaging showing distinct macular atrophy in both eyes and a choroidal naevus at left superotemporal arcade (e and f). AF imaging showing further atrophy at the central macula with central foveal sparing (g and h). Optical coherence tomography (OCT) showing preservation of the ellipsoid zone in the foveal area demarcated by red arrows, and the extent of loss of the ellipsoid zone denoted by yellow arrows (i and j)
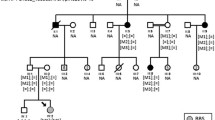
Segregation analysis in the family revealed that her father (I:2) carried the c.4685 T > C variant, but the proband’s affected sister (II:4) did not, indicating that this ABCA4 variant was not relevant to the macular dystrophy in this family (Fig. 3).
Targeted sequencing of 70 genes associated with retinal degeneration was performed in the proband [5]. A heterozygous variant, c.121C > T (p.R41W), was identified in the cone-rod homeobox (CRX) gene. Sanger sequencing confirmed the variant was present in all three affected family members (Fig. 3). This variant has previously been reported as a dominant pathogenic variant that causes cone-rod dystrophy (CRD) [6]. The family were informed of the genetic diagnosis and had follow-up genetic counselling for an autosomal dominant disease inheritance pattern associated with variable expressivity and reduced penetrance.
Discussion and conclusions
Single ABCA4 variants are frequently identified in patients phenotypically diagnosed with Stargardt macular dystrophy (STGD1) [2,3,4], ABCA4 testing was undertaken because of the family history of STGD1 and because the siblings had a bull’s eye macular dystrophy (BEM) appearance, which has been reported with ABCA4 variants [7]. However once the family survey was performed, with no ABCA4 variant identified in II/4, and a macular dystrophy diagnosed in her asymptomatic father (I:2), an ABCA4 retinopathy was excluded. The gene panel testing identified a pathogenic sequence variant in CRX. The ABCA4 variant was designated to be an incidental finding (carrier status) (Fig. 3).
The CRX (cone rod homeobox) gene was first identified as an important regulator of photoreceptor cell development by Chen et al. in 1997 [8]. Then Swain et al. and Freund et al. reported that CRX caused autosomal dominant cone rod dystrophy (CRD) [6, 9]. Swain et al. first described the c.121 T > C variant with a similar phenotype to that seen in the family we report here, except that their case in addition to rod degeneration also had cone dysfunction, which was not seen in the proband in our family [6]. A recent study by Nishiguchi et al. highlighted a range of phenotypes, observing that CRX mutations occurring downstream in the homeobox domain (p.L299F, p.G243X, p.S204fs and p.S213fs) had diffuse or posteriorly extending bone-spicule pigmentation and vascular attenuation consistent with retinitis pigmentosa whilst all their other patients with mutations in the homeobox domain had a phenotype that was consistent with a macular dysfunction or CRD typified by a central region of decreased AF surrounded by a ring of increased AF and loss of the EZ in the maculae of all the symptomatic CRX patients [10]. In addition to variable expressivity, the Nishiguchi et al. study highlighted reduced penetrance; two of the families with the CRX variant p.R41W had affected parents with a milder phenotype than their affected children. This is similar to the father I:2 in our family, who was asymptomatic when examined at the family survey. The visual acuity level in their cohort was similar to the VA seen in our 3 patients [10]. Three studies report an electronegative ERG with some CRX mutations [6, 10, 11]. Cone dystrophy alone, as seen in the proband in this family has been previously described [12] and Hull et al. describe a rod cone dystrophy phenotype associated with this same CRX c.121C > T variant [11].
Intra and inter-familial variability of expression and reduced penetrance seem to be a hallmark of CRX [10,11,12,13,14,15]. Romdhane et al. report intra-familial variability and reduced penetrance, as seen in this family and a BEM phenotype [15]. BEM associated with CRX sequence variants has been reported by a number of authors, characterised by a central decreased AF signal surrounded by a ring of increased AF and loss of the EZ on OCT imaging and foveal preservation [11, 16]. It is clear however that the presence of BEM is not pathognomic for CRX, as BEM has been described in association with sequence variants in CRX, ABCA4 and GUCY2D [16]. The spared fovea has been seen in patients with CRX mutations before, with Boulanger-Scemama et al. reporting that half of their cohort of CRX patients had a spared fovea [17].
CRX variants have been described in association with Leber congenital amaurosis and rod cone dystrophy, in addition to the typical cone rod/macular dystrophy phenotype [11].
More recently subsequent to the testing identifying the CRX variant in this case, Wollock et al. reported on the use of genome wide enrichment analysis of qualifying variants for dominant inheritance pattern diseases using the collapsed analysis approach. This type of approach reached a study wide significance for identification of CRX mutations in patients that were clinically diagnosed with STGD1, CRD, BEM, PD and MD, but were not found to have mutations in the ABCA4 gene thus highlighting the importance of using a multiple gene panel when assessing these patients [18].
In summary this family study illustrates the complexity of genetic testing in the context of phenocopies, reduced penetrance and variable expressivity. This is illustrated by the discovery of a clinical phenotype in the asymptomatic father using multimodal imaging, revisiting the original diagnosis in the sister, and showing non segregation of the ABCA4 variant, as well as identifying the pathogenic CRX variant in all three affected family members using NGS. It highlights the importance of comprehensive genetic testing and a careful family survey. If a family survey had been performed at the initial visit, the asymptomatic father would have been identified as affected, and an autosomal dominant pattern of inheritance with segregation would have excluded ABCA4 as a causative gene. However, it is not always possible to perform a family survey, which can make interpretation of genetic testing results difficult in cases where more than one variant is identified as potentially causative and where there are known phenocopies.
With the advent of whole genome analysis many potential pathogenic variants will be identified in different genes; accurate phenotyping and family studies will be key in identifying the variant(s) in causative genes, and will enable diagnosis in phenocopies and complex phenotype-genotype correlations. This case highlights the variable expression and reduced penetrance seen in CRX associated retinal dystrophies. It also emphasises the value of access to NGS genetic analysis and the importance of family studies with segregation to complement and direct genetic testing.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Abbreviations
- ABCA4:
-
ATP binding cassette subfamily A member 4
- AD:
-
Autosomal dominant
- AF:
-
Autofluorescence
- AR:
-
Autosomal recessive
- BEM:
-
Bull’s eye maculopathy
- CRD:
-
Cone-Rod dystrophy
- CRX:
-
Cone-rod homeobox
- OCT:
-
Optical coherence tomography
- STGD1:
-
Stargardt disease
References
Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, et al. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 1997;277(5333):1805–7.
Downes SM, Packham E, Cranston T, Clouston P, Seller A, Nemeth AH. Detection rate of pathogenic mutations in ABCA4 using direct sequencing: clinical and research implications. Arch Ophthalmol (Chicago, Ill: 1960). 2012;130(11):1486–90.
Zaneveld J, Siddiqui S, Li H, Wang X, Wang H, Wang K, et al. Comprehensive analysis of patients with Stargardt macular dystrophy reveals new genotype-phenotype correlations and unexpected diagnostic revisions. Genet Med. 2015;17(4):262–70.
Zernant J, Schubert C, Im KM, Burke T, Brown CM, Fishman GA, et al. Analysis of the ABCA4 gene by next-generation sequencing. Invest Ophthalmol Vis Sci. 2011;52(11):8479–87.
Shanks ME, Downes SM, Copley RR, Lise S, Broxholme J, Hudspith KA, et al. Next-generation sequencing (NGS) as a diagnostic tool for retinal degeneration reveals a much higher detection rate in early-onset disease. Eur J Hum Genet. 2013;21(3):274–80.
Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, et al. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19(6):1329–36.
Michaelides M, Chen LL, Brantley MA Jr, Andorf JL, Isaak EM, Jenkins SA, et al. ABCA4 mutations and discordant ABCA4 alleles in patients and siblings with bull's-eye maculopathy. Br J Ophthalmol. 2007;91(12):1650–5.
Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, et al. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19(5):1017–30.
Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, et al. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91(4):543–53.
Nishiguchi KM, Kunikata H, Fujita K, Hashimoto K, Koyanagi Y, Akiyama M, et al. Association of CRX genotypes and retinal phenotypes confounded by variable expressivity and electronegative electroretinogram. Clin Exp Ophthalmol. 2020;48(5):644.
Hull S, Arno G, Plagnol V, Chamney S, Russell-Eggitt I, Thompson D, et al. The phenotypic variability of retinal dystrophies associated with mutations in CRX, with report of a novel macular dystrophy phenotype. Invest Ophthalmol Vis Sci. 2014;55(10):6934–44.
Kitiratschky VB, Nagy D, Zabel T, Zrenner E, Wissinger B, Kohl S, et al. Cone and cone-rod dystrophy segregating in the same pedigree due to the same novel CRX gene mutation. Br J Ophthalmol. 2008;92(8):1086–91.
Sohocki MM, Sullivan LS, Mintz-Hittner HA, Birch D, Heckenlively JR, Freund CL, et al. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription-factor gene. Am J Hum Genet. 1998;63(5):1307–15.
Chapi M, Sabbaghi H, Suri F, Alehabib E, Rahimi-Aliabadi S, Jamali F, et al. Incomplete penetrance of CRX gene for autosomal dominant form of cone-rod dystrophy. Ophthalmic Genet. 2019;40(3):259–66.
Romdhane K, Vaclavik V, Schorderet DF, Munier FL, Viet TH. CRX-linked macular dystrophy with intrafamilial variable expressivity. Ophthalmic Genet. 2018;39(5):637–41.
Nasser F, Kurtenbach A, Kohl S, Obermaier C, Stingl K, Zrenner E. Retinal dystrophies with bull's-eye maculopathy along with negative ERGs. Doc Ophthalmol. 2019;139(1):45–57.
Boulanger-Scemama E, Mohand-Said S, El Shamieh S, Demontant V, Condroyer C, Antonio A, et al. Phenotype analysis of retinal dystrophies in light of the underlying genetic defects: application to cone and cone-rod dystrophies. Int J Mol Sci. 2019;20:19.
Wolock CJ, Stong N, Ma CJ, Nagasaki T, Lee W, Tsang SH, et al. A case-control collapsing analysis identifies retinal dystrophy genes associated with ophthalmic disease in patients with no pathogenic ABCA4 variants. Genet Med. 2019;21(10):2336–44.
Acknowledgements
We would like to thank the family, and Sophie Marlowe and Clare Arnison-Newgass, Research Coordinator and Ophthalmic Genetic Research Nurse respectively, for their help with this project.
Funding
We are grateful for funding from the Thames Valley Clinical Research Network, which enabled this study. The funder had no role in the design, collection, analysis and interpretation of the data in this study or in the writing of the manuscript. SA-K is funded by the Qatar Foundation (GSRA6-1-0329-19010).
Author information
Authors and Affiliations
Contributions
AHN and SMD initially designed the concept of this work. KAZH, SB, SH, MES and PC analysed the patient data. SA-K, KAZH, SB, SH and SMD drafted the article. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethics Committee approval was obtained from the Essex 2 Research Ethics Committee (reference 08/H0302/96) with written patient consent, and this study was conducted in adherence to the tenets of the Declaration of Helsinki.
Consent for publication
Written informed consent was obtained from the patients for this case report and any accompanying images.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Al-Khuzaei, S., Hudspith, K.A.Z., Broadgate, S. et al. Targeted next generation sequencing and family survey enable correct genetic diagnosis in CRX associated macular dystrophy – a case report. BMC Ophthalmol 21, 168 (2021). https://doi.org/10.1186/s12886-021-01919-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12886-021-01919-1