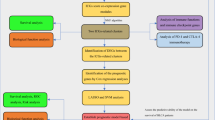
Abstract
Background
In the past few decades, researchers have made promising progress, including the development of immune checkpoint inhibitors (ICIs) in the therapy of bladder cancer (BLCA). Existing studies mainly focus on single immune checkpoint inhibitors but lack relevant studies on the gene expression profiles of multiple immune checkpoints.
Methods
RNA-sequencing profiling data and clinical information of BLCA patients and normal human bladder samples were acquired from the Cancer Genome Atlas and Gene Expression Omnibus databases and analyzed to identify different expression profiles of immune checkpoint genes (ICGs) after consensus clustering analysis. Based on the 526 intersecting differentially expressed genes, the LASSO Cox regression analysis was utilized to construct the ICG signature.
Results
According to the expression of ICGs, BLCA patients were divided into three subtypes with different phenotypic and mechanistic characteristics. Furthermore, the developed ICG signature were independent predictors of outcome in BLCA patients, and was correlated with the immune infiltration, the expression of ICGs and chemotherapeutic effect.
Conclusions
This study systematically and comprehensively analyzed the expression profile of immune checkpoint genes, and established the ICG signature to investigate the differences in ICGs expression and tumor immune microenvironment, which will help risk stratification and accelerate precision medicine. Finally, we identified KRT23 as the most critical model gene, and highlighted KRT23 as a potential target to enhance immunotherapy against BLCA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bladder cancer (BLCA) is the seventh most common type of malignant tumor worldwide [1]. Moreover, approximately 549,393 people were newly diagnosed with bladder cancer, and 199,922 patients died of the disease in 2018 [2]. Besides basic diagnosis based on symptoms and physical exams, exfoliated urothelial cells and cystoscopy are the primary methods of diagnosing bladder cancer, making BLCA one of the most expensive cancers. Approximately 80% of newly diagnosed patients present with non-muscle-invasive bladder cancer (NMIBC), and 10–20% of them will progress and invade the bladder muscle [3]. Muscular invasive bladder cancer (MIBC) is more prone to lymph node and organ metastasis, and the MIBC survival rate of five-years remains low [4]. Traditional therapy for bladder cancer mainly includes surgical resection and local or systemic chemotherapy, but the postoperative recurrence and the distant metastasis are difficult to control [5]. Therefore, it is urgent to develop new detection and treatment strategies to reduce the risk of recurrence and progression.
Although cisplatin-based combination chemotherapy realized the ideal reduction of death risk induced by bladder cancer, chemoresistance is developed and remains one of the chief reasons for cancer recurrence [6]. Additionally, the age-related and/or disease-related comorbidities may delay or impede the receipt of cisplatin-based perioperative chemotherapy [7]. As our understanding of the interaction between tumor cells and the tumor immune microenvironment (TIME) deepens, the increasing use of immune checkpoint inhibitors (ICIs) has revealed encouraging clinical efficacy in treatment. Interestingly, compared with other cancers, MIBC is one of the tumors with the highest mutation profiles, while cancers with high mutation burden, such as melanoma and non-small cell lung cancer, respond very well to ICIs [8,9,10,11]. In the past few decades, researchers had made promising progresses in the treatment of bladder cancer, including ICIs [12]. For example, a press in 2023 indicated that the combination of cisplatin/gemcitabine with nivolumab demonstrated statistically significant improvements in progression-free survival (PFS) and overall survival (OS) compared to using cisplatin/gemcitabine alone [13]. At present, the US Food and Drug Administration (FDA) has approved several ICIs for the treatment of cisplatin-ineligible patients and patients with advanced BLCA who progress despite cisplatin-based chemotherapy [14]. Despite it showed great prospects relative to past therapies, it must be recognized that only a subset of patients achieves long-term durable responses [15]. A major challenge in the ICIs therapy area is to investigate the mechanism of ICGs in BLCA and to identify patients that may respond to specific immunotherapies.
In the present study, we divided BLCA patients into three ICG clusters and built the ICG signature on the basis of the differentially expressed genes among different clusters to predict progression and outcome of BLCA. Besides, we discussed the correlation of ICG signature with the tumor immune microenvironment, and further investigated the potential mechanisms between ICGs and basic TIME characterizations. Here, our study identifies a new ICG signature to clarified the progress mechanism in bladder cancer and provides potential biomarkers and therapeutic targets.
Results
Overview of genetic changes and expression variations of immune checkpoint genes in BLCA
Among the 412 samples in TCGA-BLCA dataset, 92 samples (about 22.33%) showed ICGs mutations, and KDR showed the highest frequency of mutations (Fig. 1A). Additionally, waterfall diagram showed that missense mutation and nonsense mutation were the most common mutation types. Through the analysis of the frequency of the copy number variation (CNV) changes, we also found that most of CNV changes of ICGs were concentrated in the gain of copy number, and the gain of CD244, CD48, SLAMF1 and CD160 were the most obvious (Fig. 1B). The CNV changes of the ICGs on the chromosome were shown in Fig. 1C. In order to study the tumorigenic mechanism at the expression level, we further analyzed the expression levels of ICGs between normal and tumor samples. 30 ICGs were identified as genes with statistically differential expression finally (Fig. 1D). Those findings revealed the pathogenesis of BLCA more comprehensively, and may provide guidance for medication guide of ICIs.
Survey of genetic changes and expression variations of immune checkpoint genes in BLCA. (A) The landscape of mutation profiles of 68 ICGs in 412 bladder cancer patients from TCGA-BLCA cohort. (B) The CNV variation frequency of ICGs. red circle: amplified frequency; green circle: missing frequency. (C) The location of CNV alteration of ICGs on chromosomes. CNV, copy number variations. (D) The expression difference of 68 ICGs between normal and BLCA samples. ***p < 0.001, **p < 0.01, *p < 0.05
Tumor classification based on the ICGs
After merging the GEO and TCGA databases, we performed the survival analysis to clarify the prognostic implication of these ICGs in BLCA samples. The K-M analysis of all the prognosis related ICGs in the merged cohort was shown in Supplementary Table S1. In addition, we analyzed the correlation network containing all ICGs of prognostic functions (Fig. 2A). Based on the expression levels of 53 ICGs, we performed the consensus clustering analysis and divided all samples into three clusters to investigate the relationship between these ICGs and BLCA subtypes (Fig. 2B). The OS of three subtypes was significantly different (p < 0.05). And the survival advantage of cluster C was higher than that of cluster B and A successively (Fig. 2C). The correlation heatmap demonstrated the expression of ICGs and the clinical manifestations including age, gender, grade and TMN stage of these three different clusters (Fig. 2D). The result showed that most ICGs expressed highly in cluster C and lowly in cluster B. Then we analyzed immune atlas of ICG clusters and found that activated immune cell infiltration, including the presence of dendritic cells (DCs), macrophages, natural killer cells (NKs), neutrophil, along with activated B, CD4 and CD8 T cells, was abundant in cluster C, which typically indicates significant survival benefits (Fig. 2E). Finally, the principal component analysis indicated that patients were well divided into three clusters (Fig. 2F).
Tumor classification based on the ICGs. (A) The correlation network of all the prognosis related ICGs with correlations of absolute values greater than 0.2. Red indicates positive correlation and blue indicates negative correlation. (B) The consensus score matrix of all samples when k = 3 in merged cohorts (TCGA-BLCA, GSE13507 and GSE31684) based on the expression of ICGs. (C) OS curves for the three ICG clusters based on 670 patients with BLCA. (D) The heatmap of gene expression and clinical correlation among different clusters of BLCA. (E) Differential expression of immune cells among the BLCA ICG clusters. ***p < 0.001, **p < 0.01, *p < 0.05. (F) PCA analysis of the different ICG clusters based on the prognosis related ICGs
GSVA and functional enrichment analysis between ICG clusters
We performed GSVA to detect differential pathway activity of these three clusters and provides an intuitive and stable context for assessing biological activity (Fig. 3A and C). The result showed cluster C was significantly enriched in enrichment pathways related to complete immune activation, including CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION, T_CELL_RECEPTOR_SIGNALING_PATHWAY, B_CELL_RECEPTOR_SIGNALING_PATHWAY, CHEMOKINE_SIGNALING_PATHWAY, NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY, NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY and TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY (Fig. 3B and C). Surprisingly, compared with cluster B, cluster A was significantly enriched in innate immune cell infiltration including CHEMOKINE_SIGNALING_PATHWAY, T_CELL_RECEPTOR_SIGNALING_PATHWAY, B_CELL_RECEPTOR_SIGNALING_PATHWAY and NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY, while the prognosis of the patients in cluster A was significantly worse than that of patients in cluster B (Fig. 3A). Thus, we suspected that this may be due to the aberrant activity of REGULATION_OF_ACTIN_CYTOSKELETON and ECM_RECEPTOR_INTERACTION signaling pathway.
GSVA and Functional Enrichment Analysis Between ICG clusters. (A-C) The heatmap of GSVA enrichment analysis among different ICG clusters. Red indicates the active pathway, and blue indicates inhibition pathway. (A: cluster A vs. cluster B; B: cluster A vs. cluster C; C: cluster B vs. cluster C). (D) The Venn plot of intersection DEGs. (E) Bubble of GO enrichment analysis. (F) Bubble of the top 30 KEGG signaling pathways. BP: biological process; CC: cellular component; MF: molecular function; KEGG: Kyoto encyclopedia of genes and genomes
Subsequently, we determined DEGs among different subtypes and extracted 526 intersecting DEGs for subsequent analysis (Fig. 3D). GO enrichment analysis and KEGG pathway analysis were then performed to further explore the differences in the functions and pathways of these intersecting DEGs. BPs were mainly enriched in T cell activation, negative regulation of immune system process, leukocyte cell-cell adhesion and regulation of T cell activation. CCs were mainly enriched in external side of plasma membrane, secretory granule membrane, endocytic vesicle and MHC protein complex. MFs were mainly enriched in receptor ligand activity, signaling receptor activator activity, chemokine activity, immune receptor activity and cytokine receptor binding (Fig. 3E). KEGG were mainly enriched in cytokine − cytokine receptor interaction, phagosome, antigen processing and presentation, Th1 and Th2 cell differentiation and TNF signaling pathway (Fig. 3F). The results indicated that the intersecting DEGs were clearly correlated with the immunomodulation functions, inflammatory cell chemotaxis and epithelial mesenchymal transition (EMT) related factors.
Tumor classification based on the intersecting DEGs
The Univariate Cox regression analysis was used to analyze the prognostic values of intersecting DEGs in the merged cohort, and a total of 127 genes were considered as genes associated with prognosis (supplementary Table S2). We then performed the unsupervised clustering analysis to verify the correlation between these prognosis related intersection DEGs and BLCA subtypes (Fig. 4A). Based on the highest intragroup correlations and lowest intergroup correlations when k = 2, we classified all BLCA patients into two clusters, and we found that the prognosis of patients from cluster A was significantly better than that of the patients from cluster B (Fig. 4B). The expression of prognosis related intersection DEGs and the clinical information including age, gender, grade, TNM stage were showed in a heatmap, and we found there were significant expression differences between two clusters and little differences in clinical features (Fig. 4C). The analysis of ICGs different expression showed that the expression levels of most ICGs were significantly reduced in the cluster A, while expression of CD24, CD96, CEACAM1, TNFRSF14, TNFRSF25 and VTCN1 were increased significantly in cluster A (Fig. 4D).
Tumor classification based on the intersecting DEGs. (A) The consensus score matrix of all samples when k = 2 based on the expression of intersecting DEGs. (B) Survival curve for the two clusters. (C) Heatmap and the clinicopathologic characters of the two clusters classified by these intersecting DEGs. (D) Differential expression of ICGs among the intersecting DEGs clusters. ***p < 0.001, **p < 0.01, *p < 0.05
Establishment and validation of a ICG signature
The samples in the merged dataset were randomly assigned into training cohort and testing cohort. We performed the least absolute shrinkage and LASSO Cox regression analysis using 127 prognosis related genes in the training cohort to build the 9-gene risk signature (Fig. 5A). The regression coefficients of the 9 signature genes were shown in Table 1. The alluvial map was used to visualize the model construction and testing (Fig. 5B). Furthermore, the risk score of the patients in ICG cluster A was higher than that in ICG cluster B and ICG cluster C in succession (Fig. 5C), while the patients in genecluster B favored high-risk scores compared to those with in genecluster A (Fig. 5D). It was obvious to note that increased risk scores were shown to correlate with poor prognosis. Wilcoxon test was applied to investigate the relationship between the expression of these ICGs and BLCA risk subtypes (Fig. 5E).
Establishment of a ICG signature. (A) LASSO Cox regression analysis. (B) Alluvial diagram showing the changes of ICG clusters, intersecting DEGs clusters, the signature and fustat. (C) The ICG signature risk scores in different ICG clusters. (D) The ICG signature risk scores in different intersecting DEGs clusters. (E) Differential expression of ICGs between low- and high-risk subgroups. ***p < 0.001, **p < 0.01, *p < 0.05
We divided patients in total, training and testing cohorts into high and low risk groups with median risk score of training cohort. We found that the low group had a clear survival advantage over the high group in total (Fig. 6A), training (Fig. 6B) and testing cohorts (Fig. 6C). The subsequent ROC analysis demonstrated that this ICG signature could reliably evaluate the efficiency of prognosis prediction, and we found that AUC in training cohort was 0.777 for 1-year, 0.709 for 2-year, and 0.706 for 3-year survival (Fig. 6E). In addition, the signature was respectively validated in the total and testing cohorts (Fig. 6D and F). The risk scatterplot and heatmap revealed the difference of survival status distribution and model gene expression between low-risk and high‐risk groups in total, training, and testing cohorts (Fig. 6G and I). Moreover, we observed consistent results in the GSE13507 and GSE31684 dataset (Supplementary Fig S1). The 9 ICGs‐related genes forming the signature exhibited distinct expression diverse expression patterns, including five relatively highly expressed genes (TNFAIP6, CPVL, SLFN11, KRT23 and SPRR3) and four relatively lowly expressed genes (CD3D, GBP4, PLEKHA6 and PPP1R3C), consistent with the risk coefficient.
Validation of the ICG signature. (A-C) The KM analysis of the overall survival of the high-risk and low-risk groups in total (A), training (B) and testing cohorts (C). (D-F) The ROC analysis to evaluate the predictive efficiency of the ICG signature in total (D), training (E) and testing cohorts (F). (G-I) Distribution of the risk scores, survival status, and expression of the nine risk genes in total (G), training (H) and testing cohorts (I)
Subsequently, we conducted an in-depth analysis of differentially expressed genes between high-risk and low-risk patient groups, uncovering significant biological disparities between the two groups (Supplementary Fig S2A and S2B). Our GO analysis identified several biological processes and cellular compartments where the DEGs are enriched, including collagen fibril organization, extracellular matrix organization, collagen-containing extracellular matrix (Supplementary Fig S2C). The KEGG enrichment analysis identified significant enrichment of pathways related to cytoskeleton organization in muscle cells, ECM-receptor interaction, IL-17 signaling, and protein digestion and absorption in the DEGs (Supplementary Fig S2D). These findings suggest that the dysregulation of these pathways may contribute to the aggressive behavior of bladder cancer, including increased cell motility, altered ECM-cell interactions, immune evasion, and altered nutrient utilization. Subsequently, we constructed a PPI network of the identified DEGs to identify potential hubs and interactors (Supplementary Fig S2E). These proteins may play crucial roles in bladder cancer development and could serve as potential therapeutic targets or drug discovery leads.
Evaluation of the ICG signatureAs shown in Fig. 7A, the risk scores and all significant factors for prognosis (including age, gender, grade, TNM stage) were integrated into a novel prognostic nomogram to provide a reliable quantitative tool for predicting the prognosis of BLCA patients. The nomogram could effectively predict the 1, 3 and 5 year survival rates of BLCA patients.
Molecular characteristics and clinical utility of the ICG signature. (A) Nomograph for predicting the overall survival of BLCA patients at 1, 3, and 5 years. (B) Correlation heatmap of the levels of infiltration of the immune cells with nine risk genes. (C) Distribution of immune scores, stromal scores and ESTIMATE scores in low- and high-risk subgroups. (D-E) Differences in the mutational status within the different subgroups. (F) Expression difference of the three hot immune checkpoint genes (PD-L1, PD-1, and CTLA4) in different subgroups. (G) Box plots of differential chemosensitivity between two subgroups
A comparative analysis between the high- and low-risk groups stratified by our ICG signature revealed notable differences in tumor immune dysfunction and exclusion (TIDE), microsatellite instability (MSI), dysfunction, and exclusion scores. Specifically, the low-risk group exhibited higher TIDE and T-cell dysfunction scores, as well as a lower T-cell exclusion score, indicating a significant variation in the responsiveness to immunotherapy between the two risk groups (Supplementary Fig. 3A-3 C). However, there was no discernible difference in the MSI score between the two subgroups (Supplementary Fig. 3D). Subsequently, ROC curve analysis was employed to assess the prognostic superiority of our ICG signature model over the existing TIDE and 18-gene T-cell inflammation signature (TIS) models. The TIDE model evaluates the degree of immune cell dysfunction and exclusion within the tumor microenvironment, while the 18-gene TIS model delves into the expression of a panel of immune-related genes to predict patient prognosis [16]. Our model demonstrated an AUC value that exceeded those of both the TIDE and 18-gene TIS models (Supplementary Fig. 3E), suggesting that the ICG signature holds the potential to enhance the accuracy of risk stratification and prognosis prediction for bladder cancer patients when compared to the existing methods.
Identification of the immune characteristics and tumor mutational burden (TMB)
Correlation analysis of 9 model genes and invasive immune cell types was used to study the potential impact of ICG signature on BLCA immune microenvironment (Fig. 7B). Further investigation on the correlation between infiltrating immunocytes and risk scores revealed that risk scores had positive correlations with macrophages M0, macrophages M2, neutrophils, dendritic cells activated, T cells CD4 memory resting and mast cells activated, and risk scores had negative correlations with T cells CD8, T cells regulatory (Tregs), T cells gamma delta, Macrophages M1 and activated T cells CD4 memory (supplementary Fig S4). To quantify differences of tumor microenvironment (TME) between high and low-risk groups, ESTIMATE scores were calculated and demonstrated that high-risk group had significantly higher ESTIMATE, stromal, and immune scores compared to low-risk group (Fig. 7C).
Afterwards, we compared the differences of TMB between different risk subgroups. We found that the frequency of tumor mutation in high-risk group (95.07%) was higher than that in low-risk group (92.57%), and the most common types of mutation were missense mutation and nonsense mutation (Fig. 7D and E). Next, we determined the top 20 genes with the highest mutation rate in the risk subgroups and found that TP53 was the common mutation gene, with the mutation rates of over 40% in both risk groups.
The role of ICG signature in immunotherapy and chemotherapy
Previous studies have shown that anti PD1/PD-L1 immunotherapy is long-term effective for patients with high TMB status [17], and only a handful of patients benefit from ICI therapies. Therefore, we further explored the differential expression of most common ICIs, and found that three hot immune checkpoint genes (PD-L1, PD-1, and CTLA4) in the TCGA-BLCA cohort were up regulated in low-risk group, indicating that a better response to immunotherapy (Fig. 7F). As previously mentioned, combination chemotherapy based on cisplatin is still the first-line treatment standard for locally advanced or metastatic urothelial carcinoma [15], and methotrexate, docetaxel and paclitaxel were also shown to have significant and curative effect in bladder cancer [18,19,20]. Thus, we investigated the role of the risk model in predicting the efficacy of chemotherapy in BLCA patients. We found that high risk patients were inclined to possess lower half inhibition concentration (IC50) of chemotherapy drugs (such as cisplatin docetaxel and paclitaxel), while the sensitivity of the low-risk group to methotrexate was higher than that of the high-risk group (all p < 0.05, Fig. 7G). These results suggested that this ICG signature may help in selecting chemotherapeutic drugs and judging the therapeutic effects.
Furthermore, we profiled T cells using single-cell RNA to assess the T cell composition of the tumor environment. Series GSE149652 was downloaded from GEO database and composed of human bladder tumors and non-malignant tissue of 2 standard-of-care-untreated patients (“untreated”), 1 chemotherapy-treated patient (gemcitabine + carboplatin, “chemo”), and 4 anti-PD-L1-treated patients (“anti-PD-L1”). Unsupervised clustering of immune cell infiltration data revealed the presence of T cell components, including CD4 T, CD8 T, Tregs, and NK cells (Fig. 8A). Next, we assessed the T cell-inflamed and checkpoint gene differences in different treatment groups and found that the T cell-inflamed and checkpoint gene expressions in anti-PD-L1 group were generally higher than those in the other two groups (Fig. 8B).
T cell infiltration pattern. (A) SNE plot of total sample cells and T cell infiltration profiles of all samples. (B) T cell-inflamed and checkpoint gene differences in different treatment groups. (C and D) To further evaluate the importance of these genes for the prognostic contribution of BLCA patients, we performed a random forest analysis of these genes based on risk scores and patient survival status. (E-F) The expression difference of KRT23 in different T cell types
To further evaluate the importance of these model genes for the prognostic contribution of BLCA patients, we performed a random forest analysis of these genes based on risk scores and patient survival status and found that the mean decrease Gini of KRT23 was higher in both analyses (Fig. 8C and D). Immune infiltration analysis revealed that KRT23 expression was negatively correlated with the infiltration of CD8 + T and CD4 + cells in bladder cancer, while positively correlated with the infiltration of dendritic cells and neutrophils, indicating a complex role for KRT23 in shaping the immune microenvironment of bladder cancer (Supplementary Fig S5). Additionally, we mainly analyzed the KRT23 expression in different T cell types and found KRT23 was generally low expressed in T cells, which was similar to the results of data merged by TCGA and GEO database (Fig. 8E and F).
Knockdown of KRT23 inhibited BLCA Cell proliferation, migration and invasion, and promoted apoptosis
The expression of KRT23 increased in the normal, low grade and high grade samples in the HPA database progressively (Fig. 9A). Next, we performed RT-qPCR in 4 cell lines, consisting of 3 BLCA cell lines and 1 normal bladder epithelium cell line, and found that KRT23 has a strong expression in the bladder cancer cells (Fig. 9B). We selected T24 and J82 cells to further verify the importance of KRT23.
KRT23 Knockdown. (A) Immunohistochemical analysis of KRT23 in normal, low grade and high grade samples. (B) The gene expression levels of KRT23 were evaluated by real-time qPCR. (C) The knockdown of KRT23 by siRNA was detected by RT-PCR. (D) Cell colony formation assay. (E) CCK-8 assay. (F) Wound scratch assay. (G) Transwell Assay. (H) Flow cytometric dot plots. ***p < 0.01, **p < 0.01, *p < 0.05
To further explore the biological function of KRT23 in BLCA cells, the expression of KRT23 was knocked down in T24 and J82 cells using two different siRNAs. The efficiency of transfection of KRT23 siRNA was verified in mRNA levels (Fig. 9C). First, we examined the colony formation and proliferative capacity of BLCA cells after KRT23 knockdown. Inhibition of KRT23 expression markedly suppressed the colony formation and cell proliferation (Fig. 9D and E). Subsequently, we performed the wound healing assay scratch assay and transwell assay, and the results showed that KRT23 knockdown impaired the migration and invasion ability of the T24 and J82 cells (Fig. 9F and G). The apoptosis rate in the KRT23 knockdown groups of T24 and J82 cells was significantly higher than those in the si-NC groups (Fig. 9H). To evaluate the correlation between KRT23 and clinical outcome, we detected KRT23 expression in tumor and adjacent tissues of 6 BLCA patients using western blotting, and the results showed that KRT23 was highly expressed in tumor tissues (Fig. 10A). Those results indicated that KRT23 knockdown inhibited proliferation and promotes apoptosis, and KRT23 plays an important role in the process of promoting tumorigenesis.
Knockdown of KRT23 inhibited tumor proliferation in vivo. (A) Western blotting analysis was conducted to detect the expression of KRT23 in BLCA tissues and adjacent normal tissues (n = 6). (B) The tumor volumes were measured at the indicated time points. (C) The representative images of tumor. Tumor weight was measured after isolation from the mice. (D) IHC of PCNA and Ki67 in the tumors. ***p < 0.01, **p < 0.01, *p < 0.05. Original blots are presented in supplementary Fig S6
Knockdown of KRT23 inhibited tumor proliferation in vivo
Subsequently, we explored the impact of OGT on tumor formation in mice via a xenograft model. Control (shCtrl) or KRT23 knockdown (shKRT23) T24 cells were injected into the flanks of male nude mice. Compared to the control group, the tumors in KRT23 knockdown were significantly small and light in weight (Fig. 10B and C). At the end of this experiment, the mice were killed and their tumors were isolated. Immunohistochemistry of tumors showed that PCNA and ki67 were decreased due to the decreased expression of KRT23 (Fig. 10D), which indicated the inhibitory effect of KRT23 knockdown on tumor proliferation and was consistent with the findings in vitro.
Discussion
Despite significant advances in monitoring opportunities and unique therapeutic options, the survival of BLCA patients has not significantly changed in the past 30 years [21]. In view of the high frequency mutations identified by next-generation sequencing [22, 23], different biomarkers and therapies will be required to improve patients’ outcome in different clinical application. The effect of perioperative cisplatin-based combination chemotherapy on the survival rate of BLCA patients is still limited [24]. Currently, with the deepening of the research on immune checkpoint mechanism, immune-checkpoint inhibitors (CPIs) had shown great potential in the treatment of multiple types of malignancies and approved by FDA for first-line metastatic therapy in patients with cis-ineligible and PD-L1positive tumors (i.e., atezolizumab, pembrolizumab) and second-line therapies following failure of platinum-based chemotherapy(i.e., atezolizumab, pembrolizumab, nivolumab, avelumab and durvalumab) [24,25,26,27,28].
Based on reliable evidence from the background of metastatic urothelial carcinoma, CPIs are well tolerated and effective, and are expected to change the foundations of MIBC treatment in the near future [29, 30]. Therefore, to discover the ICGs molecular mechanisms underlying the development and progression of BLCA and to improve the prognosis and survival rate of BLCA patients, developing highly accurate, cost-effective, definitive test for diagnosis and surveillance is an urgent need.
This study systematically compared the altered expressions of ICGs between normal and BLCA samples, and analyzed the prognostic value of ICGs in BLCA. A total of 37 of 53 ICGs in merged cohort were associated with the OS of BLCA as shown by K-M survival analysis (p < 0.05). Based on the expression of ICGs, the consensus clustering analysis was performed to classify BLCA patients into three clusters. The prognosis of patients in cluster C was significantly better than that in clusters A and B. Stratified analysis of BLCA infiltrated immunocytes revealed innate immune cell infiltration, including the presence of DCs, macrophages, NKs, neutrophil, along with activated B, CD 4 and CD 8 T cells, was abundant in cluster C. It is known that resident innate immune cells provide anti-tumor activity by detecting tumor associated antigens (TAAs) and damage associated molecular patterns (DAMP), which is of vital importance in cancer immune editing [31]. DCs are the most important antigen-presenting cells, and can process TAA and DAMP and present them to cytotoxic T lymphocytes (CTLs), causing activation and subsequent tumor infiltration [32]. NK cells can regulate the immune surveillance and clearance of malignant cells, and their presence in the tumor microenvironment is related to better survival and lower progression rates of several solid tumors [31, 33]. Besides, an increasing body of evidence suggests that the density of infiltrating T cells and B cells have a positive association with longer OS or disease-free survival (DFS) [34,35,36]. Consistently, cluster C was significantly enriched in immune mechanism related pathways. Accordingly, we suspected that the infiltration of immune cells and the activation of immune pathways are important reasons for cluster C to gain survival advantage. To investigate the molecular differences among different clusters, we further identified the DEGs between the different clusters and identified 526 overlapping DEGs. Based on the results of GO and KEGG analyses, we’re convinced that ICGs could increase the activity of the immune system to promote immune-mediated elimination of tumor cells.
Existing literature mainly focuses on the study of single immune checkpoint inhibitors, but there is a lack of research on the gene expression profiles of multiple immune checkpoints. To comprehensively and systematically quantify the pattern of genetic modification of ICGs in individual tumors, we performed the LASSO and Cox analysis of the ICGs genes to establish an independent prognostic model which was well validated in the total and testing cohorts. The ICG signature was further identified as a stable and universally applicable prognostic assessment instrument for BLCA by the ROC curve analysis. Base on the ICG signature and other significant factors for prognosis, we structured nomogram to predict the 1, 3, and 5 year survival rates of BLCA patients.
In our signature, we found 5 of the 9 genes were inflammation related gene or antigen presenting related genes, functioning as significant components in inflammatory process of tumor initiation and progression. Because of the significant and strong association with the infiltrating immune cells, two genes (CD3D and GBP4) were paid special attention. Stratified analysis of BLCA immune microenvironment revealed a significant positive correlation between CD3D and GBP4 and antitumor immune cells (CD4 T cells, CD8 cells and macrophages M1) [36, 37], which was consistent with the risk coefficient of these two genes. The delta subunit of the CD3 complex encoded by CD3D gene is involved in T-cell development and signal transduction, which is essential for the initiation of antigen-specific T cell responses to pathogens, autoantigens and tumors [38]. CD3D defects could result in severe combined immunodeficiency autosomal recessive T-cell-negative/B-cell-positive/NK-cell-positive [39]. Besides, the expression of CD3D was consistently associated with both RFS and OS of breast cancer (BC), and was also found to be related to the response to neoadjuvant chemotherapy especially in estrogen receptor negative breast cancer, thus potentially identifying individuals responding to chemotherapy [40, 41]. Five members (GBP1, GBP2, GBP3, GBP4 and GBP5) of the Guanylate-binding proteins (GBPs) family are the most abundant cellular proteins in response to IFN-γ, suggesting that GBPs may be important components of the innate immunity [42, 43]. Increasing evidence suggests that TMB serves as a clinically valuable biomarker for identifying patients who may benefit from immune treatment [8]. In this study, we found that the frequency of tumor mutation in high-risk group was higher than that in low-risk group, and TP53 was the most common mutation gene. The wild-type TP53 protein plays a crucial role in cell apoptosis and cell cycle regulation following DNA damage. Cells with TP53 mutations can evade apoptosis after DNA damage and transform into cancer cells. Additionally, p53 loss or mutation in cancers can disrupt myeloid and T cell function, enabling immune evasion and advancing cancer progression [44]. The accumulation of mutant TP53 protein is considered a highly specific marker for malignant tumors [45]. Therefore, we speculate that the shorter OS and “cold” tumor microenvironment in high-risk BLCA patients may partly be attributed to the heightened TP53 mutations, which exert a specific inhibitory effect on the cancer-associated immune system. However, the mechanisms by which TP53 mutations impact the immune phenotype and prognosis of BLCA require further investigation.
Further investigation on the correlation between infiltrating immunocytes and risk scores revealed that risk scores had positive correlations with macrophages M2, and risk scores had negative correlations with Macrophages M1 and T cells CD8. The infiltrating macrophages in tumors are referred to as tumor-associated macrophages, with their activated forms primarily including two types: M1-type and M2-type macrophages. M1-type macrophages could enhance T cell effector functions through releasing pro-inflammatory signals and presenting antigens. Conversely, M2 macrophages possess immunosuppressive traits, dampening effector T cell proliferation. Consequently, the polarization of M1 macrophages towards the M2 phenotype within the tumor microenvironment plays a critical role in fostering tumor immunosuppression and enabling evasion from immune surveillance, thereby indicating a bleak outlook for cancer therapy [46]. Together, the above results indicate that patients at high risk often exhibit an immunosuppressive “cold” tumor microenvironment, elucidating their resistance to immune checkpoint therapy.
Given that high tumor mutation burden has been recognized as a predictive biomarker of immune checkpoint blocking response [47], we explored whether the developed ICG signature could stratify patients according to the sensitivity of patients to ICIs. Our findings showed that three hot immune checkpoint genes (PD-L1, PD-1, and CTLA4) were significantly downregulated in high-risk group, indicating that the signature could provide new ideas for drug design of immune checkpoints. Up to now, cisplatin-based combination chemotherapy regimens, such as methotrexate, vinblastine, doxorubicin, and cisplatin (MVAC), as well as gemcitabine and cisplatin/carboplatin (GC), remain the first-line treatment standard for locally advanced or metastatic urothelial carcinoma, but the optimal chemotherapy regimen has not been definitively established. In this scenario, immunotherapy utilizing checkpoint inhibitors is the preferred second-line option, but paclitaxel or docetaxel can be utilized as second- or later-line chemotherapy and have demonstrated significant and potentially curative effects in bladder cancer [19, 20]. In cases where patients are ineligible for immunotherapy or have received checkpoint inhibitors, docetaxel may still be considered as salvage therapy. Several Phase II trials have confirmed the effectiveness of docetaxel in treating metastatic bladder cancer [48]. Additionally, paclitaxel has shown activity as a standalone agent in urothelial cancer, achieving a 42% response rate in advanced and/or metastatic bladder cancer according to a Phase II trial [49]. Our study revealed that the ICG signature we developed could directly predict the immunotherapy and chemotherapy response, and facilitate the selection of therapy medications and assessment of treatment efficacy, which may guide more precision therapy for BLCA.
KRT23 was identified as the most critical gene by random forest analysis in this signature. As a newly discovered member of the keratin family, KRT23 belongs to the acidic type I keratins [50]. Many studies showed that KRT23 expression in tumors has been associated with increased aggressiveness and decreased patient survival in many kinds of cancer, including pancreatic cancer, colorectal carcinoma, breast cancer and hepatocellular carcinoma [51, 52]. In ovarian cancer, KRT23 upregulation promoted migration and invasion of cancer cells via EMT by regulating the TGF-β/Smad signaling pathway. Our results demonstrated that KRT23 knockdown impaired proliferation, migration and invasion, and induced the apoptosis of BLCA cells. Previous research has found that KRT23 knockdown increased the secretion of CD8 + T cell-related chemokines (CCL5, CXCL9, and CXCL10), and subsequently promoting the recruitment of CD8 + T cells [53]. Consistent with this, we observed an inverse correlation between the expression level of KRT23 and the abundance of T cell infiltration. Tumors are typically classified into two categories based on T cell infiltration within the TME: non-inflamed tumors (lacking T cell infiltration) and inflamed tumors (with T cell infiltration) [54]. Immunotherapy, including immune checkpoint inhibitors (ICIs) such as anti-PD-1, anti-PD-L1 and anti-CTLA-4 therapies, is generally ineffective in non-inflamed tumors due to the absence of T cell immunity. Consequently, inflamed tumors generally respond better to immunotherapy, including ICIs [55, 56]. In light of these discoveries, KRT23 emerges as a promising biomarker for predicting immune cell infiltration in tumors and developing novel immunotherapy strategies. However, further biological experiments are needed to clarify the regulatory mechanism and to promote the signature as new anticancer drug targets.
Conclusion
Taken together, this study systematically and comprehensively analyzed the expression profile of immune checkpoint genes in BLCA and their correlation with tumor mutation. In addition, we established the ICG signature to investigate the differences in immune checkpoint expression and tumor immune microenvironment, which will help risk stratification and accelerate precision medicine. Finally, we identified KRT23 as the most critical model gene and highlighted it as a potential target to enhance immunotherapy against BLCA.
Materials and methods
Data source and preprocessing
Overall RNA sequencing data and clinical information of BLCA patients and normal human bladder samples were obtained from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/) and GEO database (https://www.ncbi.nlm.nih.gov/geo/, ID: GSE13507 and GSE31684). The patient characteristics of the collected patients in TCGA and GEO database were shown in Supplementary Table S3. Processed single-cell RNA sequencing data came from tumor immune single-cell hub 2 (TISCH2) database (http://tisch.comp-genomics.org) based on GSE149652 data from the GEO database [57]. Moreover, somatic mutation data are downloaded from TCGA database and presented with the mutation landscape by “maftools” package. The Wilcoxon test was applied to identify differentially expressed ICGs. Then, we used univariate Cox regression analysis and Kaplan-Meier survival estimates to extract ICGs associated with prognosis using the R packages ‘survival’ and ‘survminer’ (p-value < 0.05). The correlation of ICGs were visualized by the “igraph” package in the R software (https://www.r-project.org/).
ICG clusters
We screened and identified 68 immune checkpoint genes with the key words of immune checkpoint, immune checkpoint inhibitors, immune checkpoint blockade, and checkpoint blockade in the Pubmed database (https://pubmed.ncbi.nlm.nih.gov/). All of the ICGs were shown in Supplementary Table S4. We merged BLCA samples of TCGA, GSE13507 and GSE31684 datasets into one train (n = 664) to analyze the prognostic value of the ICGs for BLCA. After merging the GEO and TCGA databases, we found that the remaining 53 ICGs were expressed in different datasets. According to 53 ICGs expression level, we performed unsupervised cluster analysis by R package “Consensus Cluster Plus” to explore the underlying molecular subtype between the BLCA patients [58]. The maximum cluster count (k) was set as 9, and the optimal cluster count value was chosen for further study. Kaplan Meier analysis was used for survival analysis and the Log rank analysis for comparison to evaluate overall survival (OS) differences with ‘survival’ package in R. Principal component analysis (PCA) was displayed to visualize the similarities and differences among grouped clusters. We followed the methods of Ninghong Song1 et al. 2022 [59].
Gene set variation analysis (GSVA)
GSVA was used to assess changes in pathways and biological process movement among different clusters. We downloaded the “c2.cp.kegg.v7.4.symbols” gene set from the molecular signatures database for GSVA. The adjusted p value less than 0.05 is considered statistically significant.
Distinguish the differentially expressed genes (DEGs) related to ICGs
The differentially expressed genes among different clusters were analyzed with “limma” package, and the venn diagram of the overlapped genes between each cluster was draw by “VennDiagram” package. The significance criteria for selecting differentially expressed genes related to ICGs was set as an adjusted p < 0.05 and the value of |logFC| > 1. To further explore the biological processes and signal pathways of the intersection genes, Gene Ontology (GO) functions of the overlapping genes and enhanced the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were analyzed with the “clusterProfiler” package.
Establishment and validation of the prognostic signature
The relationship between the intersection DEGs and survival was explored by the univariate Cox analysis. The cut-off p-value was set to 0.05, and 127 intersection genes related to survival were screened for further study. All BLCA patients (n = 664) were randomly divided to a training cohort (n = 332) (50% for identifying the prognostic model) and a testing cohort (n = 332) (50% for validating the prognostic model). The LASSO Cox regression model was utilized to minimize overfitting between survival-related genes and to obtain a more concise prognostic genes combination by “glmnet” package. Finally, we construct the 9-gene risk signature based on multivariate Cox regression, and calculated the risk score by multiplying the expression level of each gene by the corresponding regression coefficient. The BLCA samples were assigned into high- and low-risk groups according to the median risk score. The area under the ROC curve (AUC value) obtained from the R package “timeROC” was used to evaluate the diagnostic efficacy of prognostic model. The waterfall map of high- and low-risk group mutation landscape was presented by maftools function. The immunohistochemistry (IHC) staining images of risk genes in normal and BLCA tissues using the same antibodies were analyzed and obtained from the Human Protein Atlas (HPA) database (http://www.proteinatlas.org/).
Assessment of tumor immune microenvironment
CIBERSORT was applied to evaluate the immunocyte infiltration levels of BLCA, and the ESTIMATE algorithm was used to evaluate the immune scores and the stromal scores. We performed correlation analysis on the ICG signature to further reveal the connection with immune cells. In addition, the most common immune checkpoint genes, such as PD-1, PD-L1 and CTLA4, were extracted and evaluated the relevance with ICG signature in TCGA-BLCA dataset.
Drug sensitivity
Using “pRRophetic” package in R, we estimated the chemotherapeutic response determined of the common anticancer drugs by the half-maximal inhibitory concentration (IC50) and compared the difference of chemosensitivity between the high- and the low-risk group.
Cell culture, RT-PCR and siRNA transfection
The immortalized cells of human bladder epithelium (SV-HUC-1) and human BLCA cell line (T24, UMUC3 and J82) were cultured in Ham’s F-12 K (Gibco/BRL, Grand Island, NY), RPMI-1640 medium (HyClone, Logan, UT, USA) and MEM (Gibco/BRL, Grand Island, NY) supplemented with complete medium (10% fetal bovine serum) at 37 °C and 5% CO2. Total RNA was isolated using TRIzol reagent (Invitrogen, Frederick, MD, USA), and cDNA was synthesized using standard techniques. Quantitative RT-PCR analyses were performed using SYBR Green Fast qPCR Mix (ABclonal, RK21203, Wuhan, China) on Prism® 7500 (Applied Biosystems, California, USA). The KRT23 primers were 5′- CCATGCAGAATCTCAACGAC.
-3′ (sense) and 5′- GGTGTGTGATGTTTTCCTCA-3′ (antisense). The GAPDH primers were 5′- GATGGAGGAGGCTCAGCA − 3′ (sense) and 5′- CTCAGCCAATGGGACCTG − 3′ (antisense).
The sequences of KRT23-target-specifc-siRNA were as follows: siRNA1: 5′-AUGAGAAUGAGCUCUUUCCUC-3′(sense), 5′-GGAAAGAGCUCAUUCUCAUGA-3′(antisense); siRNA2:5′-GGAGGAUAUGAGACAAGAAUA-3′(sense), 5′-UUCUUGUCUCAUAUCCUCCAG-3′(antisense). The shRNA expression vector targeting KRT23 was synthesized based on the sequence of siRNA2. The INTERFERin (Polyplus, Illkirch-Graffenstaden, France) was used to transfect siRNA and vector into cells according to the manufacturer’s instructions.
Proliferation studies
2000 cells per well of T24 and J82 cell lines were seeded in 96-well plates after KRT23 siRNA transfection. The cell viability was determined at 12 h, 24 h, 48 h, 72 h and 96 h. T24 and J82 cell suspension with logarithmic growth phase was inoculated with 1000 cells/well in a 6-well plate. After 2 weeks, the cells were fixed with 4% paraformaldehyde for 20 min and stained with 0.1% crystal violet for 20 min.
Wound scratch assay and transwell assay
After transfected with KRT23 siRNA, the 200 µL eppen-dorf tip was used to scratch the cell plate. The complete medium was then replaced with serum-free medium, and cell migration was observed at 0 h and 48 h with an inverted microscope.
Migration and invasion of T24 and J82 cells was analyzed through the Transwell assay. 3 × 104 cells in medium without serum were seeded in the upper chamber of the Transwell apparatus without/with Matrigel (ABW, 0827045, Xiamen, China). Then 500 µL medium with 10% fetal bovine serum (FBS) was added to the lower chamber. The membrane of chamber was fixed for 30 min and stained with 0.1% crystal violet after 24 h.
Apoptosis assay
Apoptosis of T24 and J82 cells were detected using the Annexin V-FITC apoptosis detection kits (Elabscience, E-CK-A211, Wuhan, China). 1 × 105 cells in 500 µL1× binding buffer were labeled with Annexin V-FITC (5 µL) and propidium iodide (PI) (5 µL) for 15 min at room temperature in the dark. Specimens were analyzed with a Accuri C6 Plus flow cytometer within 1 h of labeling.
Western blotting
Western blotting was performed as previously studies described [60]. The following antibodies were used: anti-KRT23 (Abcam, ab156569, 1:1000 dilution) and anti-GAPDH (ABclonal, a19056, 1:50000 dilution).
Tumor tissue preparation
we collected six fresh bladder cancer samples and their paired adjacent normal samples from BLCA patients who underwent radical cystectomy at Beijing Chaoyang Hospital Affiliated Capital Medical University, without any preoperative treatment. The study received approval from the Medical Ethical Committees of the Beijing Chaoyang Hospital Affiliated Capital Medical University. All patients were informed about this study.
Tumor xenograft assay and immunohistochemistry staining
Six-week-old female BALB/c nude mice (Vital River Laboratories, China) were randomly divided into two groups (n = 4 per group). Next, 1 × 107 KRT23 knockdown (shKRT23 group) and negative control (shCtrl group) T24 cells were subcutaneously injected into the right flank of two groups of mice. All animals were deeply anesthetized before euthanasia to ensure they were unconscious and did not experience pain. All animals were deeply anesthetized before euthanasia to ensure they were unconscious and did not experience pain. Isoflurane was used as the anesthetic, administered through inhalation by placing the animals in an induction chamber and exposing them to 4–5% isoflurane in oxygen for 2–3 min until deep anesthesia was achieved. After confirming deep anesthesia, euthanasia was performed using cervical dislocation, a method that is quick and ensures a humane death. All experimental procedures were approved by the Animal Ethics Committee of Beijing Chaoyang Hospital, Capital Medical University.
IHC staining for PCNA (Boster, BM0104, 1:200 dilution) and ki67 (Abcam, ab16667, 1:500 dilution) was conducted following the manufacturer’s instructions as described in previous study [61].
Statistical analysis
The Student’s t test was utilized for statistical comparison of paired data, while the ANOVA test for comparison of more than two groups and Pearson’s chi-square tests for comparison of categorical variables. Logarithmic rank test was used to determine the significance of differences in survival curves. Statistical significance was set at p < 0.05.
Data availability
The datasets analyzed during the current study are available in the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/), GEO database (http://www.ncbi.nlm.nih.gov/geo, ID: GSE13507 and GSE31684), HPA database (http://www.proteinatlas.org/) and TISCH2 database (http://tisch.comp-genomics.org/home/).
Abbreviations
- BLCA:
-
Bladder cancer
- ICIs:
-
Immune checkpoint inhibitors
- ICGs:
-
Immune checkpoint genes
- NMIBC:
-
Non-muscle-invasive bladder cancer
- MIBC:
-
Muscular invasive bladder cancer
- FDA:
-
Food and Drug Administration
- TIME:
-
Tumor immune microenvironment
- PCA:
-
Principal component analysis
- GSVA:
-
Gene set variation analysis
- DEGs:
-
Differentially expressed genes
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- GO:
-
Gene Ontology
- IC50:
-
Half-maximal inhibitory concentration
- CNV:
-
Copy number variation
- GBPs:
-
Guanylate-binding proteins
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Teramoto Y, Jiang G, Goto T, Mizushima T, Nagata Y, Netto GJ, Miyamoto H. Androgen receptor signaling induces Cisplatin Resistance via Down-regulating GULP1 expression in bladder Cancer. Int J Mol Sci. 2021; 22.
Zhang S, Liu Y, Liu Z, Zhang C, Cao H, Ye Y, Wang S, Zhang Y, Xiao S, Yang P, Li J, Bai Z. Transcriptome profiling of a multiple recurrent muscle-invasive urothelial carcinoma of the bladder by deep sequencing. PLoS ONE. 2014;9:e91466.
Zhao J, Zhou L, Pan Y, Chen L. A systematic review and meta-analysis of radical cystectomy in the treatment of muscular invasive bladder cancer (MIBC). Transl Androl Urol. 2021;10:3476–85.
Powles T, Bellmunt J, Comperat E, De Santis M, Huddart R, Loriot Y, Necchi A, Valderrama BP, Ravaud A, Shariat SF, van der Szabados B, Gillessen S, et al. Bladder cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2022;33:244–58.
Jain P, Kathuria H, Momin M. Clinical therapies and nano drug delivery systems for urinary bladder cancer. Pharmacol Ther. 2021;226:107871.
Sternberg CN, Skoneczna I, Kerst JM, Albers P, Fossa SD, Agerbaek M, Dumez H, de Santis M, Theodore C, Leahy MG, Chester JD, Verbaeys A, Daugaard G, et al. Immediate versus deferred chemotherapy after radical cystectomy in patients with pT3-pT4 or N + M0 urothelial carcinoma of the bladder (EORTC 30994): an intergroup, open-label, randomised phase 3 trial. Lancet Oncol. 2015;16:76–86.
Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, Kaley TJ, Kendall SM, Motzer RJ, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–6.
Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor Mutational Burden as an independent predictor of response to Immunotherapy in Diverse Cancers. Mol Cancer Ther. 2017;16:2598–608.
Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, van den Akerley W, Lutzky J, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23.
Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, Barlesi F, Kohlhaufl M, Arrieta O, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-small-cell Lung Cancer. N Engl J Med. 2015;373:1627–39.
Chen X, Xu R, He D, Zhang Y, Chen H, Zhu Y, Cheng Y, Liu R, Zhu R, Gong L, Xiao M, Wang Z, Deng L, et al. CD8(+) T effector and immune checkpoint signatures predict prognosis and responsiveness to immunotherapy in bladder cancer. Oncogene. 2021;40:6223–34.
van Dorp J. The bladder cancer immune micro-environment in the context of response to immune checkpoint inhibition. Front Immunol. 2023;14:1235884.
Patel VG, Oh WK, Galsky MD. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J Clin. 2020;70:404–23.
Tran L, Xiao JF, Agarwal N, Duex JE, Theodorescu D. Advances in bladder cancer biology and therapy. Nat Rev Cancer. 2021;21:104–21.
Chen Y, Li ZY, Zhou GQ, Sun Y. An Immune-Related Gene Prognostic Index for Head and Neck squamous cell carcinoma. Clin Cancer Res. 2021;27:330–41.
Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. M(6)a regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19:53.
Yagoda A, Memorial Sloan-Kettering Cancer Center. Phase-II trials in patients with urothelial tract tumors. Cancer Chemother Pharmacol. 1983;11(Suppl):S9–12.
Lobo N, Mount C, Omar K, Nair R, Thurairaja R, Khan MS. Landmarks in the treatment of muscle-invasive bladder cancer. Nat Rev Urol. 2017;14:565–74.
Mitin T, Hunt D, Shipley WU, Kaufman DS, Uzzo R, Wu CL, Buyyounouski MK, Sandler H, Zietman AL. Transurethral surgery and twice-daily radiation plus paclitaxel-cisplatin or fluorouracil-cisplatin with selective bladder preservation and adjuvant chemotherapy for patients with muscle invasive bladder cancer (RTOG 0233): a randomised multicentre phase 2 trial. Lancet Oncol. 2013;14:863–72.
Abdollah F, Gandaglia G, Thuret R, Schmitges J, Tian Z, Jeldres C, Passoni NM, Briganti A, Shariat SF, Perrotte P, Montorsi F, Karakiewicz PI, Sun M. Incidence, survival and mortality rates of stage-specific bladder cancer in United States: a trend analysis. Cancer Epidemiol. 2013;37:219–25.
Sjodahl G, Lauss M, Lovgren K, Chebil G, Gudjonsson S, Veerla S, Patschan O, Aine M, Ferno M, Ringner M, Mansson W, Liedberg F, Lindgren D, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res. 2012;18:3377–86.
Seiler R, Ashab HAD, Erho N, van Rhijn BWG, Winters B, Douglas J, Van Kessel KE, van de Fransen EE, Sommerlad M, Wang NQ, Choeurng V, Gibb EA, Palmer-Aronsten B, et al. Impact of Molecular subtypes in muscle-invasive bladder Cancer on Predicting Response and Survival after Neoadjuvant Chemotherapy. Eur Urol. 2017;72:544–54.
Gust KM, Rebhan K, Resch I, Shariat SF, Necchi A. Immune checkpoint inhibition in muscle-invasive and locally advanced bladder cancer. Curr Opin Urol. 2020;30:547–56.
Apolo AB, Infante JR, Balmanoukian A, Patel MR, Wang D, Kelly K, Mega AE, Britten CD, Ravaud A, Mita AC, Safran H, Stinchcombe TE, Srdanov M, et al. Avelumab, an anti-programmed death-ligand 1 antibody, in patients with refractory metastatic urothelial carcinoma: results from a Multicenter, phase ib study. J Clin Oncol. 2017;35:2117–24.
Balar AV, Galsky MD, Rosenberg JE, Powles T, Petrylak DP, Bellmunt J, Loriot Y, Necchi A, Hoffman-Censits J, Perez-Gracia JL, van der Dawson NA, Dreicer R, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet. 2017;389:67–76.
Balar AV, Castellano D, O’Donnell PH, Grivas P, Vuky J, Powles T, Plimack ER, Hahn NM, de Wit R, Pang L, Savage MJ, Perini RF, Keefe SM, et al. First-line pembrolizumab in cisplatin-ineligible patients with locally advanced and unresectable or metastatic urothelial cancer (KEYNOTE-052): a multicentre, single-arm, phase 2 study. Lancet Oncol. 2017;18:1483–92.
Sharma P, Callahan MK, Bono P, Kim J, Spiliopoulou P, Calvo E, Pillai RN, Ott PA, de Braud F, Morse M, Le DT, Jaeger D, Chan E, et al. Nivolumab monotherapy in recurrent metastatic urothelial carcinoma (CheckMate 032): a multicentre, open-label, two-stage, multi-arm, phase 1/2 trial. Lancet Oncol. 2016;17:1590–8.
Resch I, Shariat SF, Gust KM. PD-1 and PD-L1 inhibitors after platinum-based chemotherapy or in first-line therapy in cisplatin-ineligible patients: dramatic improvement of prognosis and overall survival after decades of hopelessness in patients with metastatic urothelial cancer. Memo. 2018;11:43–6.
Gust KM, Abufaraj M, D’Andrea D, Moschini M, Soria F, Shariat SF. Second line immune checkpoint inhibition in urothelial cancer. Transl Androl Urol. 2019;8:414–20.
Daza J, Charap A, Wiklund PN, Sfakianos JP. Role of the Innate Immune System in the Development, Progression, and therapeutic response of bladder Cancer. Eur Urol Focus. 2020;6:650–2.
Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annu Rev Immunol. 2013;31:51–72.
Morrison BJ, Steel JC, Morris JC. Reduction of MHC-I expression limits T-lymphocyte-mediated killing of Cancer-initiating cells. BMC Cancer. 2018;18:469.
Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, Signoretti S, Liu JS, Liu XS. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174.
Li XD, Huang CW, Liu ZF, Jiang LJ, Chen JW, Xie D, Zhou FJ, Lu HM, Liu ZW. Prognostic role of the immunoscore for patients with Urothelial Carcinoma of the bladder who underwent Radical Cystectomy. Ann Surg Oncol. 2019;26:4148–56.
van Wilpe S, van der Gerretsen ECF, de Vries IJM, Gerritsen WR, Mehra N. Prognostic and predictive value of Tumor-infiltrating Immune cells in Urothelial Cancer of the bladder. Cancers (Basel). 2020; 12.
Komohara Y, Jinushi M, Takeya M. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 2014;105:1–8.
Kuhns MS, Davis MM, Garcia KC. Deconstructing the form and function of the TCR/CD3 complex. Immunity. 2006;24:133–9.
Gil J, Busto EM, Garcillan B, Chean C, Garcia-Rodriguez MC, Diaz-Alderete A, Navarro J, Reine J, Mencia A, Gurbindo D, Belendez C, Gordillo I, Duchniewicz M, et al. A leaky mutation in CD3D differentially affects alphabeta and gammadelta T cells and leads to a Talphabeta-Tgammadelta + B + NK + human SCID. J Clin Invest. 2011;121:3872–6.
West NR, Milne K, Truong PT, Macpherson N, Nelson BH, Watson PH. Tumor-infiltrating lymphocytes predict response to anthracycline-based chemotherapy in estrogen receptor-negative breast cancer. Breast Cancer Res. 2011;13:R126.
Gyorffy B, Bottai G, Fleischer T, Munkacsy G, Budczies J, Paladini L, Borresen-Dale AL, Kristensen VN, Santarpia L. Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. Int J Cancer. 2016;138:87–97.
Cheng YS, Colonno RJ, Yin FH. Interferon induction of fibroblast proteins with guanylate binding activity. J Biol Chem. 1983;258:7746–50.
Britzen-Laurent N, Bauer M, Berton V, Fischer N, Syguda A, Reipschlager S, Naschberger E, Herrmann C, Sturzl M. Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS ONE. 2010;5:e14246.
Blagih J, Buck MD, Vousden KH. p53, cancer and the immune response. J Cell Sci. 2020; 133.
Long J, Wang A, Bai Y, Lin J, Yang X, Wang D, Yang X, Jiang Y, Zhao H. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine. 2019;42:363–74.
Guo B, Yang F, Zhang L, Zhao Q, Wang W, Yin L, Chen D, Wang M, Han S, Xiao H, Xing N. Cuproptosis Induced by ROS responsive nanoparticles with Elesclomol and Copper Combined with alphaPD-L1 for enhanced Cancer Immunotherapy. Adv Mater. 2023;35:e2212267.
Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, Peters S. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44–56.
Albany C, Sonpavde G. Docetaxel for the treatment of bladder cancer. Expert Opin Investig Drugs. 2015;24:1657–64.
Lu Z, Yeh TK, Wang J, Chen L, Lyness G, Xin Y, Wientjes MG, Bergdall V, Couto G, Alvarez-Berger F, Kosarek CE, Au JL. Paclitaxel gelatin nanoparticles for intravesical bladder cancer therapy. J Urol. 2011;185:1478–83.
Zhou J, Qian W, Huang C, Mai C, Lai Y, Lin Z, Lai G. Combined targeting of KRT23 and NCCRP1 as a potential novel therapeutic approach for the treatment of triple-negative breast cancer. Gland Surg. 2022;11:1673–82.
Qin Y, Chen L, Chen L. Identification and verification of key cancer genes associated with prognosis of colorectal cancer based on bioinformatics analysis. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2021;46:1063–70.
Gao X, Yang J. Identification of genes related to clinicopathological characteristics and prognosis of patients with colorectal Cancer. DNA Cell Biol. 2020;39:690–9.
Li X, Cheng Y, Cheng Y, Shi H. Transcriptome analysis reveals the Immune Infiltration profiles in Cervical Cancer and identifies KRT23 as an immunotherapeutic target. Front Oncol. 2022;12:779356.
Gajewski TF. The next hurdle in Cancer Immunotherapy: overcoming the Non-t-cell-inflamed Tumor Microenvironment. Semin Oncol. 2015;42:663–71.
Zemek RM, De Jong E, Chin WL, Schuster IS, Fear VS, Casey TH, Forbes C, Dart SJ, Leslie C, Zaitouny A, Small M, Boon L, Forrest ARR et al. Sensitization to immune checkpoint blockade through activation of a STAT1/NK axis in the tumor microenvironment. Sci Transl Med. 2019; 11.
Li H, Chen J, Li Z, Chen M, Ou Z, Mo M, Wang R, Tong S, Liu P, Cai Z, Zhang C, Liu Z, Deng D, et al. S100A5 attenuates efficiency of Anti-PD-L1/PD-1 immunotherapy by inhibiting CD8(+) T cell-mediated anti-cancer immunity in bladder carcinoma. Adv Sci (Weinh). 2023;10:e2300110.
Oh DY, Kwek SS, Raju SS, Li T, McCarthy E, Chow E, Aran D, Ilano A, Pai CS, Rancan C, Allaire K, Burra A, Sun Y et al. Intratumoral CD4(+) T Cells Mediate Anti-tumor Cytotoxicity in Human Bladder Cancer. Cell. 2020; 181: 1612-25 e13.
Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3.
Zhang X, Wei X, Wang Y, Wang S, Ji C, Yao L, Song N. Pyroptosis regulators and Tumor Microenvironment Infiltration characterization in Clear Cell Renal Cell Carcinoma. Front Oncol. 2021;11:774279.
Zhong B, Cheng B, Huang X, Xiao Q, Niu Z, Chen YF, Yu Q, Wang W, Wu XJ. Colorectal cancer-associated fibroblasts promote metastasis by up-regulating LRG1 through stromal IL-6/STAT3 signaling. Cell Death Dis. 2021;13:16.
Zhao S, Guo W, Li J, Yu W, Guo T, Deng W, Gu C. High expression of Y-box-binding protein 1 correlates with poor prognosis and early recurrence in patients with small invasive lung adenocarcinoma. Onco Targets Ther. 2016;9:2683–92.
Acknowledgements
Everybody who made a contribution to the research has been listed as an author.
Funding
None.
Author information
Authors and Affiliations
Contributions
WW and HZC conceived of and directed the project. CDS, ZX and CHY designed the study. CDS and CHY performed the experiments. WHJ, ZX and CDS collected and analyzed the data. All the authors listed have approved the manuscript that is enclosed for publication.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Medical Ethical Committees of Beijing Chaoyang Hospital Affiliated Capital Medical University for human research (approval number 2024-2-21-1), with informed consent obtained from all participating patients. Additionally, the Animal Ethical Committee provided approval for the animal research component (approval number AEEI-2023-275), ensuring ethical standards were upheld across the study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, D., Cao, H., Zheng, X. et al. Immune checkpoint gene signature assesses immune infiltration profiles in bladder cancer and identifies KRT23 as an immunotherapeutic target. BMC Cancer 24, 1024 (2024). https://doi.org/10.1186/s12885-024-12790-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-024-12790-w